Key resources
Key resources for this work, including novel plasmids deposited to Addgene, are listed in Supplementary Table 2.
Plasmid DNA
Standard molecular cloning techniques were used to generate DNA constructs in this study. Double-stranded DNA was synthesized by Integrated DNA Technologies and inserted into pAAV backbones with NEBuilder HiFi (New England Biolabs, E2621). sgRNA sequences were synthesized as overlapping single-stranded DNA oligos (Integrated DNA Technologies) that were then annealed together and ligated into sgRNA expression cassettes using T4 DNA ligase (New England Biolabs, M0202). Plasmids used in Supplementary Data Fig. 2 were constructed from polymerase chain reaction (PCR)-amplified DNA fragments (Integrated DNA Technologies) assembled via Golden Gate Assembly (New England Biolabs, E1602S). Sequences of sgRNAs and utilized DNA elements (for example, promoters and enhancers) are provided in Supplementary Table 3. Self-complementary pAAVs were generated from pscAAV-CAG-GFP, a gift from Mark Kay (Addgene, 83279).
pUCmini-iCAP-AAV-PHP.eB13 (Addgene, 103005), pUCmini-iCAP-AAV.CAP-B10 (ref. 15) (Addgene, 175004), pUCmini-iCAP-AAV.MaCPNS2 (ref. 16) (Addgene, 185137), AAV-DJ rep-cap (Cell Biolabs, VPK-420-DJ), AAV6 rep-cap (Cell Biolabs, VPK-426) and pHelper (Agilent Technologies, 240071) plasmids were used for production of AAVs. Before use, all plasmids were sequence verified via whole-plasmid sequencing through Plasmidsaurus using Oxford Nanopore Technology with custom analysis and annotation.
AAV production
Detailed protocols for AAV production and titration are available on protocols.io (https://doi.org/10.17504/protocols.io.n2bvjnew5gk5/v1 and https://doi.org/10.17504/protocols.io.e6nvw1n47lmk/v1). AAVs were produced and purified according to published methods77, with some minor alterations. In brief, HEK293T cells (American Type Culture Collection (ATCC), CRL-3216; RRID: CVCL_0063) were triple transfected with PEI-MAX (Polysciences, 24765) to deliver the rep-cap or iCAP, pHelper and genome packaging plasmids. Viruses were harvested from cells and media and then purified over 15%, 25%, 40% and 60% iodixanol (OptiPrep; Serumwerk, 1893) step gradients. A Type 70 Ti fixed-angle titanium rotor (Beckman Coulter, 337922) at 58,400 r.p.m. for 1.5 h or a Type 70.1 Ti fixed-angle titanium rotor (Beckman Coulter, 342184) at 61,700 r.p.m. for 1.25 h was used, depending on the scale and number of AAVs to be purified simultaneously. Viruses were concentrated using Amicon Ultra-15 or Amicon Ultra-4 filters with a 100-kD size cutoff (MilliporeSigma, UFC9100 and UFC8100) and formulated in sterile DPBS (Thermo Fisher Scientific, 14190144) with 0.001% Pluronic F-68 (Thermo Fisher Scientific, 24040032). AAVs were titered with quantitative PCR (qPCR) by measuring the number of DNase I-resistant viral genomes, relative to a linearized genome plasmid standard. Before injection, AAVs were diluted in sterile saline. Sequences of qPCR primers for titering are provided in Supplementary Table 3.
ssAAV genomes were used for all experiments, except those shown in the following, where scAAV genomes were used: Fig. 2c and Extended Data Fig. 4a–c; Fig. 2d and Extended Data Fig. 4d,e; Extended Data Fig. 5; Fig. 3f,g and Extended Data Fig. 6; as well as indicated parts of Fig. 2b and Supplementary Data Fig. 1.
Tissue culture
For AAV production, and for some in vitro experiments, HEK293T cells were used (ATCC, CRL-3216; RRID: CVCL_0063). Cells were grown in DMEM (Thermo Fisher Scientific, 10569010) supplemented with 10% defined FBS (Cytiva, SH30070.03).
For small-scale HEK293T experiments, cells were seeded at optimal confluence (50% for transduction, 90% for transfection) in the morning and transfected or transduced in the afternoon. For transfection, Lipofectamine LTX (Thermo Fisher Scientific, 15338100) was used, with 500 ng of total of DNA and 3 μl of transfection reagent. To avoid saturating SpECTr or fluorescent protein signal, 50 ng of DNA (for Extended Data Fig. 7) or 1,000 double-stranded DNA copies per cell (for Supplementary Data Fig. 2) was used, with pUC19 (New England Biolabs, N3041S; RRID: Addgene_50005) used as filler to ensure efficient transfection. For investigation of transcriptional crosstalk with transfection and transduction in vitro (Extended Data Fig. 7), we transduced cells with a 1 × 105 MOI of AAV-DJ, and cells were collected 5 d later. For in situ restriction enzyme digest of AAV concatemers (Fig. 3c–e and Extended Data Fig. 5), an MOI of 1 × 106 AAV-DJ was used, and cells were collected 3 d later. On the morning of collection, we passaged cells 1:10 onto poly-d-lysine-coated coverslips (Neuvitro, GG-12-1.5h-PDL). Once HEK293T cells had attached, the coverslips were washed three times in DPBS and then fixed. For analysis of fluorescent protein expression, cells were fixed with ice-cold 4% paraformaldehyde (PFA; Electron Microscopy Sciences, 15714-S) in 1× PBS for 15 min at 4 °C and stored in 1× PBS at 4 °C until use. For AAV-Zombie or SpECTr, cells were fixed with ice-cold 3:1 methanol:acetic acid (MAA; Sigma-Aldrich, 322415 and A6283) for 15 min at −20 °C and then stored at −20 °C in 70% ethanol until use.
A detailed protocol for mouse primary cortical and hippocampal neuron culture preparation is available on protocols.io (https://doi.org/10.17504/protocols.io.8epv52925v1b/v1). For primary neuron cultures, coverslips (Neuvitro, GG-12-1.5h-pre) were prepared by coating with poly-d-lysine (0.1 mg ml−1 overnight; Sigma-Aldrich, P6407), poly-l-ornithine (0.01% overnight; Sigma-Aldrich, P4957) and laminin (0.02 mg ml−1 overnight; Thermo Fisher Scientific, 23017015). Primary neurons were prepared by pooling cortices and hippocampi from several embryonic day (E) 16.5 embryos and digesting the tissue in 15 U ml−1 papain (Sigma-Aldrich, P3125). The cell suspension was then treated with DNase I, and cells were triturated in HBSS (Thermo Fisher Scientific, 14025092), with 5% horse serum (Thermo Fisher Scientific, 16050130), and then centrifuged through 4% BSA. The cell pellet was resuspended in NeuroCult Neuronal Plating Medium (STEMCELL Technologies, 5713), supplemented with 1:50 NeuroCult SM1 (STEMCELL Technologies, 05711), 0.5 mM GlutaMAX (Thermo Fisher Scientific, 35050061) and 3.7 μg ml−1 L-glutamic acid (Sigma-Aldrich, 49449), and plated at a density of 60,000 cells per coverslip. At 5 days in vitro (DIV), half the media was exchanged for BrainPhys Neuronal Media (STEMCELL Technologies, 05790), also supplemented with 1:50 NeuroCult SM1. For transduction, AAV was diluted in the added growth media. The removed plating media were saved and combined 1:1 with complete BrainPhys media. To minimize prolonged transduction due to AAVs in culture media, we used the 1:1 mix of conditioned plating media and BrainPhys media to perform a complete media change at 3 h after transduction, with three washes in pre-warmed BrainPhys between the aspiration of the virus-containing media and addition of fresh conditioned media. Subsequently, the media were half-changed with supplemented BrainPhys media every 3 d. Primary neurons were harvested and fixed as described for HEK293T cells above.
Animals
Animal husbandry and all procedures involving animals were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and approved by the Institutional Animal Care and Use Committee and by the Office of Laboratory Animal Resources at the California Institute of Technology.
Eight-week old male C57BL/6J (strain no. 000664; RRID: IMSR_JAX:000664), C57BL/6J-background Prkdcscid/scid (strain no. 001913; RRID: IMSR_JAX:001913) and C57BL/6J-background Rosa26CAG-LSL-tdTomato (strain no. 007914; RRID: IMSR_JAX:007914) mice were obtained from The Jackson Laboratory. Mice were housed 3–4 per cage, on a 12-h light–dark cycle, and had ad libitum access to food and water. For behavioral experiments and EEG recordings, animals were kept in a reverse light cycle; all behavioral assays and recordings were conducted during the dark cycle, between the hours of Zeitgeber time (ZT) 13 and ZT 17.
For most animal experiments, mice were 8.5–9.5 weeks old at the start of experiments (that is, at injection or at the start of behavioral experiments). However, for EEG experiments, the animals were 8.5–12.5 weeks old at the time of surgery, as they needed to be staggered to accommodate the large cohort size. In this case, care was taken to ensure no systemic assignment to experimental groups based on age at experiment onset.
For primary neuron cultures, timed pregnant C57BL/6N (RRID: MGI:2159965) dams were obtained from Charles River Laboratories.
Retro-orbital injection
A detailed protocol for systemic AAV administration through retro-orbital injection is available on protocols.io (https://doi.org/10.17504/protocols.io.36wgqnw73gk5/v1). AAVs were administered via retro-orbital injection during isoflurane anesthesia (1–3% in 95% O2/5% CO2, provided by nose cone at 1 L min−1), followed by administration of 1–2 drops of 0.5% proparacaine to the corneal surface77.
EEG implantation surgery
A detailed protocol for mouse EEG implantation surgery and EEG data collection is available on protocols.io (https://doi.org/10.17504/protocols.io.81wgbzj2ygpk/v1). Mice were anesthetized with isoflurane (5% induction, 1% maintenance) and then subcutaneously injected with ketoprofen (5 mg kg−1) and buprenorphine XR (3.25 mg kg−1). The animals’ heads were fixed in a stereotaxic frame (David Kopf Instruments), with a heating pad to maintain body temperature. The scalp was then sterilized and subcutaneously injected with 1–2 drops of 0.5% bupivacaine, and a 1.5-cm anterior–posterior incision was made to expose the skull. The skull surface was scored with a scalpel, and the EEG headmount (Pinnacle Technology, 8201) was glued to the surface of the skull using cyanoacrylate adhesive. The anterior edge of the headmount was targeted to be 3.5 mm anterior to bregma. A sterile 23-gauge needle was used to pierce the skull underneath each hole in the headmount. EEG screws were implanted through the headmount and into the craniotomy hole; 0.10-inch screws (Pinnacle Technology, 8209) were used for the anterior holes, and 0.12-inch EEG screws (Pinnacle Technology, 8212) were used for the posterior holes. A small amount of silver epoxy (Pinnacle Technology, 8226) was applied to each screw before fully tightening to ensure electrical connection between the screw and the headmount. Continuity of the contacts was assessed with a multimeter. Adhesive cement (C&B Metabond; Parkell, S398, S371 and S396) was used to secure screws and the headmount in place, followed by dental cement to cover the edges of the headmount. Ibuprofen (30 mg kg−1) was provided in drinking water for at least 3 d after surgery. Animals were allowed to recover for at least 1 week before EEG recordings.
Tissue harvest and processing
Tissue was collected 4 weeks after AAV administration, except for animals used in the Cacna1a knockout experiments in which tissue was collected 6 weeks (for motor behavior cohort) or 8 weeks (for EEG cohort) after AAV administration. Animals were euthanized via intraperitoneal injection of 100 mg kg−1 euthasol.
Details of tissue harvest protocols for AAV-Zombie or SpECTr experiments in tissue can be found on protocols.io (https://doi.org/10.17504/protocols.io.14egn6k7yl5d/v1). In brief, animals were transcardially perfused with 30 ml of ice-cold heparinized 1× PBS, and liver and brain were dissected out. For analysis of fluorescent protein expression, one hemisphere of brain and one lobe of liver were submerged in ice-cold 4% PFA formulated in 1× PBS and fixed overnight at 4 °C. The other hemisphere and another lobe of liver were manually dissected into 1-mm3 pieces with regions of interest and flash frozen in optimal cutting temperature (OCT) compound (Scigen, 4586) using a dry ice/ethanol bath. OCT blocks were kept at −70 °C until sectioning.
For measurement of viral genomes from bulk DNA, tissue was processed as above, except that unfixed tissue was used immediately for genomic DNA extraction (DNeasy Blood and Tissue Kit; Qiagen, 69504) rather than frozen.
If animals were not used for AAV-Zombie, SpECTr or bulk DNA extraction, then, after perfusion with PBS, animals were perfused with 30 ml of ice-cold 4% PFA in 1× PBS. Relevant tissues were then extracted and post-fixed overnight in 4% PFA in 1× PBS at 4 °C. For sectioning, brain and liver were cryoprotected through immersion in 30% sucrose in 1× PBS. Once the tissue had sunk, it was flash frozen in OCT compound using a dry ice/ethanol bath and kept at −70 °C until sectioning.
Sections were obtained using a cryostat (Leica Biosystems). Fixed tissue was sectioned at 80 μm, collected in 1× PBS and stored at 4 °C until use. Tissue for AAV-Zombie or SpECTr was sectioned at 20 μm, collected on a clean glass slide (Brain Research Laboratories, 2575-plus), allowed to dry and then stored at −70 °C until use.
Immediately before imaging, gut tissue and DRG were optically cleared by overnight room temperature incubation in RIMS78,79 and then mounted in RIMS with an iSpacer (SunJin Lab). Gut tissue was cut longitudinally before incubation in RIMS and mounted with the myenteric plexus up.
Digital droplet PCR
A detailed protocol for quantification of AAV genomes from total DNA with digital droplet PCR (ddPCR) is available on protocols.io (https://doi.org/10.17504/protocols.io.8epv5r84dg1b/v1). To measure viral genomes from bulk cortex and liver DNA, ddPCR was used. First, 1 μg of total DNA was digested overnight with 20 U of SmaI (New England Biolabs, R0141) at 25 °C or with 20 U each of KpnI-HF and SpeI-HF (New England Biolabs, R3142 and R3133) at 37 °C. The digests were diluted 1:10, and 5 μl of each dilution was loaded into a 25-μl PCR reaction (Bio-Rad, 1863024). Then, 23 μl of the PCR reaction was used to generate droplets (Bio-Rad, 1863005) on a QX200 Droplet Generator (Bio-Rad). Forty microliters of droplets was transferred to a PCR plate, which was sealed with a pierceable heat seal (Bio-Rad, 1814040 and 1814000), and the PCR was run according to the manufacturer’s protocol. After PCR, droplets were measured with a QX200 Droplet Reader and analyzed using QX Manager software (Bio-Rad, 12010213). Double-quenched FAM-labeled and HEX-labeled probe assays (Integrated DNA Technologies) were used to detect EGFP sequence and W3SL sequence in the same droplets, and the mean of the two resultant concentrations was used. Sequences of ddPCR primer and probe sets are provided in Supplementary Table 3. SmaI and KpnI-HF/SpeI-HF digests yielded similar results; only SmaI digests are shown.
IHC
A detailed protocol for IHC on mouse brain slices is available on protocols.io (https://doi.org/10.17504/protocols.io.5qpvokmq7l4o/v1). IHC, except against Cacna1a, was performed on free-floating sections. For IHC detection of Cacna1a, sections were first mounted onto slides and subjected to heat-induced epitope retrieval by boiling in 1× citrate buffer, pH 6 (Sigma-Aldrich, C9999) for 10 min in a microwave, followed by thorough washing with 1× PBS.
For IHC, sections were blocked in BlockAid Blocking Solution (Thermo Fisher Scientific, B10710) with 0.1% Triton X-100 (Sigma-Aldrich, 93443). Primary and secondary antibodies were diluted in this blocking buffer. Tissue was incubated with primary antibody overnight at 4 °C and with secondary antibody for 2 h at room temperature. After each antibody incubation step, sections were washed three times for 10 min each in 1× PBS with 0.1% Triton X-100. For Hoechst labeling, sections were incubated for 10 min with 1/10,000 Hoechst 33342 (Thermo Fisher Scientific, H3570) in 1× PBS, followed by three washes in 1× PBS. For segmentation of PCs, sections were Nissl stained with 1/50 NeuroTrace 435/455 (Thermo Fisher Scientific, N21479) in 1× PBS, followed by two 1-h room temperature washes and one overnight wash at 4 °C in 1× PBS with 0.1% Triton X-100. Sections were allowed to dry on slides, and then a coverslip was mounted using Prolong Diamond Antifade Mountant (Thermo Fisher Scientific, P36965).
The following primary antibodies and dilutions were used: rabbit anti-Cacna1a (1:100; Alomone Labs, ACC-001; RRID:AB_2039764), chicken anti-GFP (1:1,000; Aves Labs, 1020; RRID:AB_10000240) and rabbit anti-TagRFP (for detection of mRuby2; 1:1,000; a generous gift from Dawen Cai, University of Michigan; Cancer Tools, 155266; RRID: AB_3107169). Fluorophore-conjugated F(ab′)2 fragment secondary antibodies (Jackson ImmunoResearch) were used at a 1:1,000 working concentration.
AAV-Zombie and SpECTr of cultured cells
A detailed protocol for AAV-Zombie and SpECTr on cultured cells is available on protocols.io (https://doi.org/10.17504/protocols.io.36wgqnz53gk5/v3). AAV-Zombie and SpECTr protocols and sequences of Zombie barcodes and their split initiator probes were adapted from Askary et al.45. Split initiator probes against endogenous genes and reporter transcripts were designed according to Jang et al.19. Sequences of HCR-FISH probes against reporter and endogenous transcripts and against Zombie/SpECTr barcodes are provided in Supplementary Table 3.
For detection of ssAAV and scAAV genomes in cell-free conditions (Supplementary Data Fig. 1), we embedded packaged AAVs (AAV-DJ serotype) in high-concentration Matrigel (Corning, 354262). AAVs were first diluted in ice-cold 1× PBS, and 30 μl of that dilution was added to a pre-chilled tube with 270 μl of high-concentration Matrigel. After mixing by pipetting and brief vortexing, 100 μl of this suspension was spread onto a PDL-coated coverslip, in a 24-well plate on ice. After gelation for 30 min at 37 °C, the samples were incubated for 15 min in ice-cold 1× PBS at 4 °C or in MAA at −20 °C.
For AAV-Zombie and SpECTr of Matrigel-embedded AAV samples and of cultured cells on coverslips, a humidified reaction chamber consisting of a 1-ml pipette tip box filled with pre-warmed RNase-free water was used. Parafilm placed on the wafer of the box served as a surface for the in situ transcription reaction. Coverslips, previously fixed in MAA and stored in 70% ethanol, were first washed twice in 1× PBS. Then, 20 μl of transcription mixture per coverslip was prepared according to the manufacturer’s protocol (Thermo Fisher Scientific, AM1334 and AM1330). For simultaneous T7 and SP6 reactions, the T7 buffer was used with 1 μl of each RNA polymerase. For single polymerase reactions, 2 μl of the polymerase was used. Twenty-microliter droplets were pipetted onto the surface of the parafilm. The coverslips were dipped in UltraPure water (Thermo Fisher Scientific, 10977015), quickly dried by touching their edges to a Kimwipe and then placed cell-side down over the droplets. This reaction was incubated at 37 °C for 3 h.
Once the transcription reaction was finished, the coverslips were placed cell-side up into a clean 24-well plate and fixed for 20 min at 4 °C with ice-cold PFA in 1× PBS. This was followed by two 5-min washes in 1× PBS, followed by two 5-min washes in 5× SSC (Thermo Fisher Scientific, AM9770). Samples were then incubated for 15–30 min in pre-warmed probe hybridization buffer, consisting of 2× SSC, 10% ethylene carbonate (Sigma-Aldrich, E26258) and 10% dextran sulfate (Sigma-Aldrich, 3730) at 37 °C. After this incubation, the coverslips were incubated for 12–16 h at 37 °C in hybridization buffer plus 2 nM of each probe. Probes for Zombie barcodes, reporter transcripts and endogenous transcripts were pooled.
After probe hybridization, samples were washed twice for 30 min in stringent wash buffer (2× SSC, 30% ethylene carbonate) at 37 °C and then three times for 15 min in 5× SSC with 0.1% Tween 20 (Sigma-Aldrich, P1379) and then incubated in HCR amplification buffer (2× SSC, 10% ethylene carbonate) for 20–30 min. HCR hairpins (Molecular Technologies) were heated to 95 °C for 90 s and then cooled to room temperature for 30 min in the dark. For HCR on cultured cells, 30 nM hairpin in amplification buffer was used in a 1-h amplification reaction. The samples were then washed four times in 5× SSC with 0.1% Tween 20 (10 min per wash, at room temperature).
In some cases, the cytoplasm was labeled with a fluorophore-conjugated poly(dT30) probe (Integrated DNA Technologies). Coverslips were incubated with 100 nM poly(dT30) probe in 5× SSC with 0.1% Tween 20 for 1 h, followed by four 10-min, room temperature washes in 5× SSC with 0.1% Tween 20. Finally, Hoechst 33342 was used to label cell nuclei. Samples were mounted with Prolong Diamond Antifade Mountant.
For co-detection of AAV genomes and capsids, a mouse anti-AAV VP1/VP2/VP3 monoclonal antibody conjugated to Alexa Fluor 488 was used (Clone B1; Progen, 61058-488; RRID: AB_3107170). After poly(dT) labeling, the samples were immunolabeled as described above, with an overnight 4 °C incubation with a 1:100 dilution of the primary antibody in blocking buffer.
For in situ restriction enzyme digest, coverslips were treated with restriction enzymes after MAA fixation and before in situ transcription. Restriction enzyme digests were carried out overnight, at 25 °C for SmaI (New England Biolabs, R0141) and at 37 °C for MscI (New England Biolabs, R0534) and BstEII-HF (New England Biolabs, R3162).
AAV-Zombie and SpECTr of tissue sections
A detailed protocol for AAV-Zombie and SpECTr on tissue sections is available on protocols.io (https://doi.org/10.17504/protocols.io.14egn6k7yl5d/v1). AAV-Zombie and SpECTr were performed on tissue sections as described above for cultured cells, save for a few differences. Incubations in tissue were performed in a staining tray (Simport, M918), and fixation and washes were done in Coplin jars.
Sliced fresh tissue was first removed from −70 °C storage and allowed to warm to room temperature. Slides were then briefly washed with 1× PBS to remove OCT compound and then fixed for 3 h in MAA at −20 °C. Residual fixative was washed off with 1× PBS while the transcription mix was prepared. A total of 200 μl of transcription mix was used per slide, which was pipetted onto the slide and spread out with a clean glass coverslip. We found that simultaneous T7 and SP6 transcription in tissue yielded relatively few and small spots from the SP6-driven barcode. Thus, we carried out T7 and SP6 transcription reactions on separate slides. Likewise, T7 RNA polymerase was used at a 1:10 dilution, whereas SP6 RNA polymerase was used at a 1:5 dilution. As with cultured cells, in situ transcription was carried out at 37 °C for 3 h.
For the HCR-FISH steps on tissue sections, we used 4 nM of each probe in an overnight 37 °C hybridization. The HCR hairpin concentration was also doubled to 60 nM. Short HCR incubations may result in low signal for endogenous transcripts, whereas long incubations can yield large, unresolvable Zombie barcode spots. Thus, we did an overnight incubation with only hairpins for endogenous transcripts and then switched the amplification solution to one containing all hairpins for 1 h.
Controls for AAV-Zombie and SpECTr
Guidelines for designing, imaging and analyzing AAV-Zombie and SpECTr experiments are available on protocols.io (https://doi.org/10.17504/protocols.io.n2bvjn72pgk5/v1). Both AAV-Zombie and SpECTr can produce signals due to hybridization of probes directly to single-stranded AAV genomes and/or transcriptional activity of the AAV ITRs producing barcoded transcripts (for example, faint ‘concatemer’ signal in Genome A condition; Fig. 3b). Thus, controls are necessary for setting thresholds for determining real versus artifactual signal. A non-transduced/non-transfected control sample was used for all AAV-Zombie and SpECTr experiments. For SpECTr experiments, a barcode-only control was used to define signal from probe hybridizing to the AAV genome and/or barcoded transcripts produced due to transcriptional activity of the ITR. As the transcriptional activity of the AAV ITR may differ between cell types, these control experiments were performed in each cell and tissue of interest and processed side by side with experimental samples to mitigate assay-to-assay variability. Depending on the needs of the experiment, other controls may have been included and are outlined in the description of those experiments.
Imaging
For imaging of fluorescent protein expression in cultured cells and for obtaining whole section images of mouse brain and liver, a Keyence BZ-X710 epifluorescence microscope was used, with a ×10, 0.45 numerical aperture (NA) air objective.
For all other imaging, a Zeiss LSM 880 was used. Imaging of fluorescent protein expression and IHC-stained tissue was accomplished with a ×10, 0.45 NA air objective. Imaging of AAV-Zombie and SpECTr signal in Matrigel, cultured cells and in tissue was performed with a ×63, 1.4 NA oil immersion objective. Imaging settings were chosen to capture full dynamic range of the signal without saturating pixels. When possible, laser power was adjusted before adjusting detector gain. Imaging settings were first optimized on control samples, before imaging of experimental samples. Fields of view were chosen while imaging non-experimental channels (for example, Hoechst or Nissl).
Image analysis for fluorescent protein expression and IHC
For all cell and nuclear segmentation, except segmentation of PCs, Cellpose80 (version 3.0.7; https://www.cellpose.org/; RRID: SCR_021716) was used. Images were batch processed using napari81 (version 0.4.19.post1; https://napari.org/stable/; RRID: SCR_022765) and the serialcellpose plugin (version 0.2.2; https://www.napari-hub.org/plugins/napari-serialcellpose). An Anaconda (version 2.5.4; https://www.anaconda.com/; RRID: SCR_025572) distribution of Python (version 3.10.14; https://www.python.org/; RRID: SCR_008394) was used. For HEK293T cells, masks were generated from phase-contrast images. For images of cortex, the fluorescent protein signal was used to generate masks.
PC bodies were segmented manually using the Fiji82 distribution of ImageJ (version 1.54f; https://fiji.sc/; RRID: SCR_002285), from images of Nissl-stained tissue (Extended Data Fig. 1d). The large size and intense Nissl staining of the PC body, relative to neighboring cells, was used to identify PCs.
For analysis of fluorescent protein intensity in HEK293T cells, cortical cells and PCs, CellProfiler83 (version 4.2.5; https://cellprofiler.org/; RRID: SCR_007358) was used. Classification of cortical cells and PCs as fluorescent protein (XFP) positive or XFP negative was also done using CellProfiler. For PCs, we determined the threshold for using empirically determined thresholds based on negative control tissue. For classification of PCs as mRuby2 positive or mRuby2 negative (Figs. 1d and 4b), a threshold of 25.5 a.u. was used, based on measured intensity of mRuby2 signal in WT animals injected with 3 × 1011 vg of only mDLX-minBG-CI-mRuby2 (Fig. 4a,b). For classification of cortical cells as mRuby2 positive or mRuby2 negative (Fig. 4f,h and Extended Data Fig. 3b,c), a threshold of 19.125 a.u. was used, based on measured intensity of segmented cortical cells from the mRuby2 channel for ‘no enhancer’ control animals (Fig. 1f). As Cellpose reliably did not detect GFP cells in the ‘no enhancer’ condition (Fig. 1f), no threshold was necessary for classification of cortical cells as EGFP positive or EGFP negative (Fig. 4f,h). The same threshold was used for all relevant experiments and was measured in animals injected with the lowest dose of the relevant AAV, to provide the most stringent threshold. For these analyses of cortical cells and PCs, three planes (850 μm × 850 μm) from at least four non-adjacent sagittal sections were quantified (that is, at least 12 volumes per animal).
Bulk protein quantification of SCID and WT mice was performed using Fiji, from three non-adjacent 100-μm sections per tissue per animal. Cortex and cerebellum were manually segmented from sagittal sections; liver sections were analyzed whole.
To quantify CRISPR–Cas9 editing of the Ai14 locus, tdTomato-positive cells were manually counted using Fiji. Three volumes (850 μm × 850 μm × 64 μm) were captured from each of at least four non-adjacent sections per animal. PCs and non-PCs were differentiated based on distinct cell morphology and location.
For analysis of Cacna1a expression in cerebellum, Fiji was also used. Four maximum intensity projections of 850 μm × 850 μm × 30 μm volumes were analyzed per animal. In each image, the molecular layer (ML) and the granular layer (GL) were manually segmented, and the total average fluorescence intensity was measured in those regions. For each image, the ML intensity was divided by the GL intensity, and then a per-animal average was determined.
Image analysis for AAV-Zombie and SpECTr
Guidelines for designing, imaging and analyzing AAV-Zombie and SpECTr experiments are available on protocols.io (https://doi.org/10.17504/protocols.io.n2bvjn72pgk5/v1). For analysis of AAV-Zombie and SpECTr spots, segmentation was performed as described above. For primary neurons and HEK293T cells, cell body masks were generated from poly(dT)-TAMRA signal and nuclear masks from Hoechst signal. PC nuclei were manually segmented in Fiji, using large nucleus size, euchromatic nuclear staining and the presence of Itpr1 transcript to positively identify PCs.
Quantification and measurement of AAV genomes and concatemers in PCs was accomplished using CellProfiler. Genome and concatemer spots were identified within segmented nuclear masks, using empirically determined spot size thresholds and robust background intensity thresholding, chosen due to the sparse foreground signal.
AAV genomes, concatemers and capsid puncta were identified in primary neurons and HEK293T cells as described above, with some exceptions. For both HEK293T cells and primary neurons, masks were size filtered, using empirically determined thresholds. Primary neuron masks were further filtered for presence of an overlapping nuclear mask, and a cytoplasmic mask was generated by subtracting the nuclear mask from the cell body mask. EGFP transcript intensity was measured in the entire cell body mask; AAV genome, concatemer and capsid puncta were quantified in both cytoplasm and nucleus. For HEK293T cells, only nuclear AAV genomes and concatemers were measured.
Animal behavior
Detailed protocols for the following behavioral assays are available on protocols.io (https://doi.org/10.17504/protocols.io.6qpvr8jbzlmk/v2). On each day of behavioral training and data collection, animals were acclimated to the testing room for at least 30 min before measurements were taken. Animals were trained on beam crossing and gait measurement assays 1–2 weeks before experimental measurements started. Behavior equipment was disinfected and deodorized between each animal or, in the case of the open field test, between each cage.
The open field apparatus consisted of four square arenas (27 cm × 27 cm), with a camera (EverFocus, EQ700) placed 1.83 m above the floor of the arenas. An EthoVision XT (Noldus, version 17.5; https://www.noldus.com/ethovision-xt; RRID: SCR_000441) was used to capture and subsequently analyze animal locomotion. Each trial consisted of a 2-min habituation period followed by a 10-min test period. To avoid confounds due to odors from non-cagemates, only animals from the same cage were recorded simultaneously. The average velocity over the course of the experimental period was determined.
The inverted wire hang test was used to measure limb strength84. Animals were placed onto a wire mesh screen (6 mm × 6 mm mesh), which was then inverted over the top of a 45-cm-tall cylinder with clean bedding in the bottom. A blinded experimenter recorded the latency to fall within a maximum trial period of 120 s. Three trials were recorded, and the average of those three trials was used.
To measure skilled locomotion using the narrowing beam assay, a clear plexiglass beam consisting of three 25-cm segments (widths 3.5 cm, 2.5 cm and 1.5 cm) was elevated above the table surface using empty clean cages, according to published protocol85. At the narrow end, an empty cage was placed on its side, and bedding from the animal’s home cage was placed inside. A white light was also placed over the broad end to motivate animals to move across the beam. For each trial, animals were placed at the end of the widest segment, with all four limbs touching the beam surface. Each trial was recorded with a video camera placed to the side and perpendicular to the beam’s length, affording a view of both left and right hindlimbs. A trial was considered complete once the animal had traversed the beam, without turning around, and entered the goal cage. Once an animal had completed three trials, the session was completed. For each trial, a blinded experimenter measured the animal’s time to cross the beam (ignoring time spent paused), and assigned a neurological score86: (7) traverses the beam successfully, with no more than four foot slips and does not grip the side of the beam; (6) traverses the beam successfully, using hindlimbs to aid in more than 50% of strides; (5) traverses the beam successfully, using hindlimbs to aid in less than 50% of strides; (4) traverses the beam successfully, using a hindlimb at least once to push forward but without bearing load on limb; (3) traverses beam successfully, by dragging hindlimbs without using them to push forward; (2) moves at least one body length but fails to traverse beam in the 120-s trial period or falls off; and (1) fails to traverse beam or falls off and does not move more than one body length. The average score and traversal time of the three trials was used for data presentation and statistics.
For gait analysis, we used MouseWalker (https://github.com/MouseWalker/MouseWalker/tree/v1), according to published protocols for hardware design and analysis87,88. A clear acrylic platform, 80 cm long, with a 5.3-cm corridor flanked by 12.5-cm-high walls was used. LED lights positioned around the platform enable tracking of animal contacts with the platform surface, through frustrated total internal reflection (fTIR) that is captured using a camera (Apple, iPhone 12 Pro) positioned under the platform. Mice were placed on one end of the corridor, and fTIR was recorded as the animal moved across the platform. Animals were recorded until they had completed three continuously moving traversals of the field of view. Data were analyzed using MouseWalker, and the resulting paw and body tracking was manually inspected by a blinded experimenter to ensure accuracy. In some cases, trials were excluded due to poor tracking.
EEG recording and analysis
A detailed protocol for mouse EEG implantation surgery and EEG data collection is available on protocols.io (https://doi.org/10.17504/protocols.io.81wgbzj2ygpk/v1). EEG recordings were conducted in clear Plexiglas cylinders (25 cm wide, 30 cm high) with ad libitum water. Mice were connected to a pre-amplifier (100× gain, 0.5-Hz high-pass EEG filter; Pinnacle Technology, 8208-SL), which was attached to a commutator (Pinnacle Technology, 8204). Data were acquired by Sirenia Acquisition (Pinnacle Technology, version 2.2.12; https://www.pinnaclet.com/software.html; RRID: SCR_016183), using a Pinnacle data conditioning and acquisition system (Pinnacle Technology, 8206), at a sampling rate of 400 Hz.
Mice were first habituated to the chamber for one session, at least 1 d before recordings began. For each session, a minimum of 90 min was recorded; only the last 60 min were analyzed. To assess ethosuximide blockade of absence seizures, mice were recorded for 90 min and then received a single intraperitoneal injection of ethosuximide (200 mg kg−1 in sterile saline; Sigma-Aldrich, E7138) and then were recorded for another 90 min. Only the last 60 min of the pre-ethosuximide and post-ethosuximide recordings were analyzed. Ethosuximide blockade experiments were performed 8 weeks after AAV injection.
EEG signal was analyzed using Sirenia Seizure Pro (Pinnacle Technology, version 2.2.13; https://www.pinnaclet.com/software.html; RRID: SCR_016184). The raw EEG signal was first bandpass filtered (1–25 Hz). A sliding window (0.8 s wide, 0.4-s increments) was used to automatically detect absence seizures using the following criteria: a root mean square (RMS) power exceeding 50 μV in the 5–8-Hz band and a mean amplitude at least two-fold higher than the baseline defined during the pre-injection recording session.
Statistics and reproducibility
Several biological replicates for each experiment are included in the corresponding figure legends. No data were excluded from analyses, except for gait analysis trials in which paw or body tracking was determined by a blinded experimenter to be inaccurate. For all violin plots, the middle dashed line is the median, and the upper and lower dashed lines are quartiles. Statistical analysis was performed with GraphPad Prism (version 10.0.3, GraphPad Software; RRID: SCR_002798) as described in the figure legends. Where relevant, all tests were two-tailed and corrected for multiple comparisons to maintain an experiment-wide alpha of 0.05.
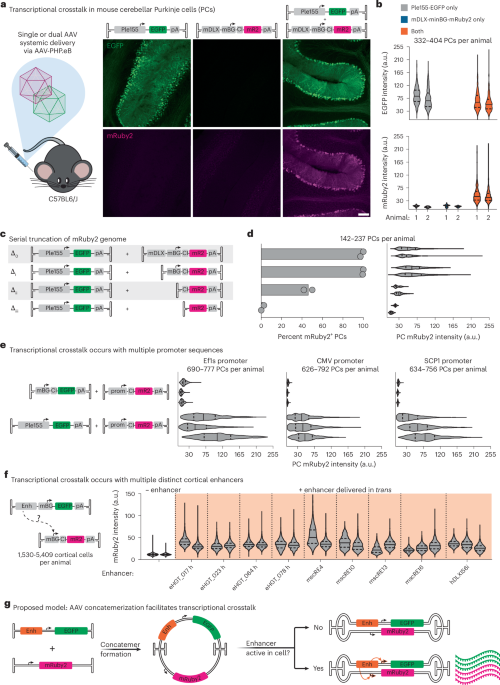
The following in vivo experiments were repeated once (n > 2 animals per experimental condition) with similar results: Fig. 1a,b and Extended Data Fig. 1a; Fig. 2d and Extended Data Fig. 4d,e; Fig. 4a–d and Extended Data Fig. 8a–e; Fig. 5a–c; Fig. 6b and Extended Data Fig. 9a,b; and Supplementary Data Fig. 3a–c. The remaining in vivo experiments were not independently repeated. All in vitro experiments were repeated at least twice with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.