Synthesis and characterization of 1,2-dicarbonyl compounds 1a–d, glutathione S-conjugates 4c, 4d and 12 and MMF
Details of the synthesis and full compound characterization are presented in the Supplementary Methods. In brief, 1,2-dicarbonyl 1a was prepared from cinnamaldehyde 6 (Supplementary Fig. 1b) in three steps. α-Keto amide 1b was synthesized as reported previously25 (Supplementary Fig. 1a). α-Ketoester 1c was synthesized from dimethyl 2-oxoglutarate 5b, using the similar bromination and elimination conditions described for 1b (Supplementary Fig. 1a)25. Phosphate buffer-mediated hydrolysis of 1c gave α-ketoacid 1d (Supplementary Fig. 1a). Glutathione S-conjugates 4c and 4d were synthesized from 1c and 1d, respectively, upon reaction with glutathione in a mixture of water and acetonitrile (Supplementary Fig. 1c). Reaction of 4d with aqueous hydrogen peroxide gave compound 12 (Supplementary Fig. 1d), a conjugate of glutathione with MMF. MMF was synthesized from maleic anhydride as reported previously26 (Supplementary Fig. 1e). Compound 1a and MMF were stored at ambient temperature with protection from light and compounds 1b–d, 4c, 4d and 12 were stored as solids below 4 °C in the absence of light.
Reaction of 1c with hydrogen peroxide
Analytical reverse-phase high performance liquid chromatography (HPLC) spectra of MMF, 1c and the reactions of 1c with hydrogen peroxide were acquired on an Agilent 1100 Infinity series machine using a Phenomenex Luna Omega PS C18 column (250 × 4.6 mm, 100 Å, 5 μm) with a buffer system of 0.1% trifluoroacetic acid (TFA) in water (buffer A) and 0.1% TFA in acetonitrile (buffer B). Elution was conducted with a gradient of 0–100% A–B over 15 min, using a flow rate of 1.0 ml min−1. To a solution of 2,778 µM, 611 µM or 5.56 µM hydrogen peroxide in water (900 µl) was added a 5 mM solution of 1c in water (100 µl) to give a final concentration of 500 µM 1c and 2,500 µM (5 equivalents), 550 µM (1.1 equivalents) or 5 µM (0.01 equivalents) hydrogen peroxide. After 40 and 130 min, the samples were analyzed using HPLC.
Compound 1c was reacted with hydrogen peroxide and the rate of MMF formation was measured by NMR spectroscopy of a sample of 1c (4.30 mg, 0.025 mmol, 1.0 equivalent) and 2,2-dimethyl-2-silapentane-5-sulfonate-d6 sodium salt (DSS-d6) (1.12 mg, 0.005 mmol) in a mixture of 111 mM phosphate buffer (pH 7.4; 450 µl) and D2O (50 µl). Then, 30% w/w hydrogen peroxide solution (5.11 µl, 0.050 mmol, 2.0 equivalents) was added to initiate the reaction. 1H NMR spectra of 1c in 100 mM phosphate buffer (pH 7.4) or water and MMF (synthesized by the authors) in 100 mM phosphate buffer (pH 7.4) were taken at a concentration of 50 mM 1c or MMF and referenced with DSS.
Reaction of 1d with glutathione
First, 100 µl each of stock solutions of 1d (20 mM) and glutathione (40 mM) in 100 mM phosphate buffer (pH 6.0, 7.0, 7.4 and 8.0) were combined in a 96-well plate to give 200 μl of reaction mixture containing 10 mM 1d and 20 mM glutathione. The absorbance at 279 nm was monitored over time using a BioTek Synergy H4 Hybrid plate reader and the resulting data were normalized to the absorbance of a 10 mM solution of 1d at the respective pH. Pseudo first-order rate constants, k (min−1), were calculated as the exponents of exponential least-squares regression functions.
Reaction of 4d with N-acetylcysteine
A 1 mM solution of 4d (0.2 ml, 0.0002 mmol) and a 1 mM solution of N-acetylcysteine (0.2 ml, 0.0002 mmol) were added to water (0.6 ml). The count of the [M + H]+ ions for each species was measured at 0.5 h and 16 h by electrospray ionization MS in positive mode on a Bruker HCT MS instrument.
In vitro NRF2 reporter assay
NRF2/ARE luciferase reporter HEK293 cells (SL-0042-NP, Signosis) were maintained in DMEM (SH30243.01, Cytiva) supplemented with 10% FBS (03-600-511, Thermo Fisher Scientific) and 1% penicillin–streptomycin (SV30010, Cytiva). Cells were seeded into 48-well flat-bottom cell culture plate (3548, Corning) in 500 μl of supplemented DMEM at a concentration of 2 × 105 cells per ml and incubated overnight at 37 °C with 5% CO2 in a humidified environment. Before treatments, the medium was replaced with fresh 0.1% FBS in DMEM. tert-Butylhydroquinone (0–30 μM; 112941, Millipore Sigma), a well-known NRF2 activator, was used as a positive control to confirm NRF2-dependent luciferase production (Supplementary Fig. 11).
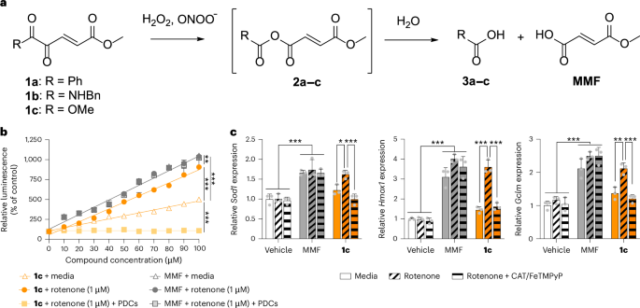
Cells were treated with a concentration range of rotenone (mitochondrial complex I inhibitor; R8875, Millipore Sigma) (0–100 μM) to identify the concentrations that did not activate NRF2 per se and were not cytotoxic (Supplementary Fig. 12). In separate experiments, cells were treated with concentration ranges of 1c or MMF (651419, Millipore Sigma) (1–100 μM) followed by rotenone (1 μM) to endogenously overproduce peroxides27. The medium was used as the negative control for all conditions.
To identify concentrations of hydrogen peroxide and peroxynitrite that did not activate NRF2 per se, cells were treated with concentration ranges of hydrogen peroxide (H1009, Millipore Sigma) or peroxynitrite (20-107, Millipore Sigma) (0–300 μM) (Supplementary Fig. 11). In separate experiments, cells were treated with concentration ranges of 1a–c, 5b (349631, Millipore Sigma) or MMF (651419, Millipore Sigma) (1–100 μM)7,8,28, followed by a fixed concentration of exogenous hydrogen peroxide (1 or 10 μM) or peroxynitrite (1 or 20 μM). These peroxide concentrations represent those in the physiological and pathological range, respectively29,30. The medium alone was used as the negative control for all conditions.
For experiments to eliminate peroxides, cells were treated with a concentration range of 1c (0–100 μM), together with 250 U per ml catalase (E3289, Millipore Sigma)31 and 50 μM FeTMPyP (75854, Cayman Chemical)32, followed by rotenone (1 μM).
Cells were incubated with treatments for 4 h for studies of gene expression or 16 h for luciferase reporter activity. To quantify luciferase activity, cells were washed with PBS (Thermo Fisher Scientific) and then lysed by a 15-min incubation at room temperature with passive lysis buffer (E1941, Promega). Cell lysates (30 μl) were transferred to a 96-well white or clear flat-bottom plate (3632, Corning) and mixed with 150 μl of luciferase substrate (LUC100, Signosis). The plates were read in a Synergy HTX multimode reader (BioTek). All conditions were performed in triplicate and observations were verified across three independent experiments.
Animals
Pathogen-free adult male and female C57Bl/6J (8 weeks old on arrival; strain 000664) and C57Bl/6J diet-induced obese (DIO) (8 weeks old on arrival; strain 380050) mice were purchased from The Jackson Laboratory (Bar Harbor). Male and female (8–12 weeks old) Nfe2l2−/− mice and wild-type littermates on a C57BL/6J genetic background (The Jackson Laboratory; strain 017009) were bred at The University of Texas MD Anderson Cancer Center. Male Sprague–Dawley rats were purchased from Inotiv. Mice were housed 5 per cage and rats were housed 3–4 per cage in a light-controlled and temperature-controlled room (12-h light–dark cycle; lights on at 7:00 a.m.) with food and water available ad libitum. Equal numbers of male and female rodents were used in each experimental group, except for pharmacokinetic studies (males only). Animals were randomly assigned to experimental groups using a randomization calculator (GraphPad Software). All animals were acclimated to the animal care facility for at least 7 days before the start of the study. Procedures were approved by the Animal Care and Use Committees of MD Anderson Cancer Center and Texas Southern University.
Surgical PNI
Spared nerve injury33 was performed in mice34. Briefly, under inhaled isoflurane anesthesia, the tibial and common peroneal nerves were isolated, tightly ligated with 6–0 silk (707G, Ethicon), and transected immediately distal to the ligation. The sural nerve was left intact. For sham surgery, the nerves were exposed but not ligated or transected. Animals were monitored postoperatively until fully ambulatory before return to their home cage.
Surgical destabilization of the medial meniscus (DMM)
DMM was performed to induce osteoarthritis35. Mice were anesthetized with isoflurane and placed in dorsal recumbency. Carprofen (5 mg kg−1, subcutaneously (s.c.)) was administered immediately before surgery began. A 3-mm longitudinal incision was made over the right patella and the joint capsule was opened with micro Vannas scissors under ×4 magnification. Blunt dissection of the infrapatellar fat pad exposed the meniscotibial ligament of the medial meniscus, which was then transected using a no. 11 scalpel blade. The incision was closed with two 9-mm AutoClip wound clips, which were removed 10 days after surgery. Animals were monitored postoperatively until fully ambulatory before return to their home cage.
CIPN
Cisplatin (TEVA Pharmaceuticals) was diluted in sterile saline and administered for 5 days (2.3 mg kg−1 day−1, intraperitoneally (i.p.)) followed by 5 days of rest and a second round of five doses to induce CIPN36.
High-fat diet (HFD)-induced neuropathy
Upon arrival, DIO mice were fed an HFD (Research Diets D12492 (60% kcal from fat) to induce type 2 diabetic neuropathy37. Mice were maintained on an HFD for the duration of the study, with drug dosing beginning 2 weeks after arrival.
In vivo drug treatments
Compound 1c (molecular weight (MW) = 172.14) and DRF (MW = 255.23) were suspended in methylcellulose (viscosity: 15 cP, 2% w/v in water; Millipore Sigma). Compound 4d (MW = 465.44) and MMF (MW = 130.10) were dissolved in 0.9% saline. For pharmacokinetic assessments, 1c (350 μmol kg−1, orally (p.o.)) was administered as a single dose. For assays of glutathione levels, mice were treated daily with 1c (350 μmol kg−1 day−1, p.o.), DRF (350 μmol kg−1 day−1, p.o.) or methylcellulose vehicle for 3 days, beginning 7 days after PNI. For thermal imaging, 7 days after PNI, mice were treated with a single dose of 1c (350 μmol kg−1 day−1, p.o.), DRF (350 μmol kg−1 day−1, p.o.) or methylcellulose vehicle.
For PNI antinociception, 1c (350 μmol kg−1 day−1, p.o.), DRF (350 μmol kg−1 day−1, p.o.) or vehicle control (equal volume, p.o.) was administered once daily for 7 days, beginning 180 days after PNI. For other PNI antinociception and postmortem tissue analyses, 1c (100, 225 or 350 μmol kg−1 day−1, p.o.), 5b (350 μmol kg−1 day−1, p.o.), DRF (350 μmol kg−1 day−1, p.o.), 4d (190 μmol kg−1 day−1, intravenously (i.v.)), MMF (190 μmol kg−1 day−1, i.v.) or an equal volume of vehicle control (p.o. or i.v., as appropriate) was administered once daily, beginning 7 days after PNI or sham surgery and continuing for 3 days (postmortem studies and reflex tests) or 7 days (conditioned place preference tests). For other postmortem tissue analyses after peroxide decomposition, catalase (250 U, i.t.)38 and FeTMPyP (5 mg kg−1, i.p.)39 or the vehicle control was administered 30 min before and 60 min after once-daily 1c (350 μmol kg−1 day−1, p.o.), DRF (350 μmol kg−1 day−1, p.o.) or vehicle control (equal volume, p.o.), beginning 7 days after PNI or sham surgery and continuing until day 9.
For DMM antinociception, 1c (350 μmol kg−1 day−1, p.o.) or methylcellulose vehicle was administered for five consecutive days, beginning 4 weeks after surgery. For antinociceptive tolerance, 1c (350 μmol kg−1 day−1, p.o.) or the positive control morphine sulfate (3 mg kg−1 b.i.d., s.c.; gifted from the National Institute on Drug Abuse Drug Supply Program, Research Triangle Institute) was administered daily for 5 days, beginning 7 days after PNI. For acute morphine analgesia, morphine sulfate (5 mg kg−1, s.c.) or saline vehicle was administered once in a randomized crossover design between 19 and 22 days after PNI. For CIPN antinociception, 1c (350 μmol kg−1 day−1, p.o.) or vehicle was administered for five consecutive days, beginning 3 days after the last cisplatin dose. For HFD-induced neuropathy antinociception, 1c (225 μmol kg−1 day−1, p.o.) or vehicle was administered for five consecutive days, beginning 4 weeks after initiation of the HFD.
In vivo pharmacokinetics
To facilitate the withdrawal of multiple, timed blood draws for pharmacokinetic analysis, the right jugular vein of each rat was cannulated 1 day before drug administration. Under anesthesia (ketamine, acetopromazine and xylazine, 50:3.3:3.3 mg kg−1, i.p.), silicone elastomer tubing (0.02 × 0.037 inches) was inserted into the jugular vein, secured with a silk suture and exteriorized in the dorsal infrascapular region. The surgical incision was prophylactically treated with nitrofurazone wound powder and closed with surgical staples. The cannulae were flushed daily with 0.5 ml of sterile heparinized saline (100 U per ml). All studies were initiated between 8:00 a.m. and 9:00 a.m. to eliminate possible circadian variation. The animals were fasted overnight and until 4 h after dosing, with water available ad libitum. Blood samples (0.15 ml) were collected from the cannulae immediately before administration of 1c and at 0.083, 0.25, 0.5, 0.75, 1, 1.5, 2, 3, 4, 6 and 8 h after dosing. Plasma samples were isolated, immediately mixed with formic acid (1:1 v/v), stored at −80 °C and analyzed within 1 week using LC–MS/MS. Concentrations were calculated from standard curves of each analyte in rat plasma.
Tissue collection
Within 4 h of the final dose, mice were deeply anesthetized with Beuthanasia-D (Merck) and then transcardially perfused with ice-cold saline. In some experiments, blood was collected by cardiac puncture before perfusion. The ipsilateral and contralateral sciatic nerve (5 mm proximal to the transection), L4 and L5 DRG, heart, liver, kidney and lung were isolated and rapidly frozen for subsequent analysis.
qPCR
Total RNA was extracted from HEK293 cells or DRG tissues using TRIzol (Thermo Fisher Scientific). Then, 1 μg of RNA was used for reverse transcription with iScript reverse transcription supermix (Bio-Rad). Real-time PCR was carried out in a final volume of 20 μl with iTaq Universal SYBR green supermix (Bio-Rad) containing 2 μl of fivefold diluted complementary DNA and monitored by a CFX Connect real-time PCR detection system (Bio-Rad). The following cycling parameters were used: 95 °C for 3 min, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Primer sequences are reported in Supplementary Table 4. The level of the target mRNA was quantified relative to the housekeeping gene (Gapdh) using the ΔΔCt method. Gapdh was not significantly different between treatments.
Western blotting
DRG or sciatic nerves from two or three mice were pooled within groups (treatment, lateralization and sex) to ensure that sufficient protein could be obtained for analysis. Liver, kidney, heart and lung were not pooled. Nuclear fractions were isolated with an NE-PER nuclear and cytoplasmic extraction kit (78835, Thermo Fisher Scientific), according to the manufacturer’s instructions. Western blotting was performed as previously described21. Nuclear proteins were subjected to NuPAGE Bis-Tris (4–12%) gel electrophoresis under reducing conditions. After transfer to nitrocellulose membranes (IB23001, Invitrogen), nonspecific binding sites were blocked with Superblock buffer (37515, Thermo Fisher Scientific) for 1 h at room temperature. Membranes were incubated overnight at 4 °C with primary anti-NRF2 antibody (1:1,000; rabbit polyclonal IgG; ab31163, Abcam) and anti-histone H3 antibody (1:2,000; rabbit polyclonal IgG; ab1791, Abcam) (loading control). The membranes were then washed with PBS containing 0.1% Tween-20 and probed with horseradish peroxidase-conjugated secondary antibody (1:5,000; goat polyclonal IgG; Jackson ImmunoResearch) in blocking buffer containing 0.1% Tween-20 for 1 h at room temperature. After washing with 1× PBS containing 0.1% Tween-20, membranes were developed with enhanced chemiluminescence substrate (Thermo Fisher Scientific). Images were acquired using ImageQuant LAS 4000 (GE Healthcare Life Sciences). Densitometry analysis was performed using ImageQuant TL software (GE Healthcare Life Sciences). Data were normalized to the loading control (histone H3) and then to the vehicle control within each gel.
Glutathione assay
Serum samples were assayed in duplicate for glutathione content (703002, Cayman Chemical), according to the manufacturer’s instructions. The plates were read in a Synergy HTX multimode reader (BioTek).
Thermal imaging
For thermographic analysis of ear skin, mice were housed individually in a small plexiglass enclosure on a mesh stand. Images were acquired between 10:30 and 11:30 a.m. using a thermal camera (C5; Teledyne FLIR), before and 30 min after treatment. The intensity of a single pixel was calculated at the center of the spot meter (Thermal Studio, Teledyne FLIR), obtained within the antihelix of the pinna (region of interest). Three measurements were made in each ear (where possible) and averaged to obtain the skin temperature for each mouse. Data are presented as the difference between the post-treatment and baseline measurements for each mouse. Image acquisition and analysis were performed by an experimenter who was blinded to treatment conditions.
Behavioral sensory testing
All behavioral tests were conducted by an experimenter who was blinded to group assignments. Mice received at least three 60-min habituations to the test environment before reflex testing. Rodents were placed in a small plexiglass enclosure on a mesh stand. Punctate allodynia was measured using the von Frey test21. The 50% paw withdrawal threshold was determined using the ‘up–down’ method40. Dose-dependent reversal of allodynia is reported as a percentage of the maximum possible antiallodynia, calculated for each mouse as a ratio of its actual antiallodynia compared to a hypothetical situation in which the drug brought withdrawal thresholds to their original baseline at the postinjection time point.
Dynamic allodynia was measured by lightly stroking the plantar surface of the hindpaw with a soft paintbrush41. A paintbrush (5/0, Princeton Art and Brush) was prepared by blunting the tip and removing the outer layer of hairs. The lateral plantar region of the left hindpaw (sural nerve territory) was stimulated by light stroking (∼2 cm s−1) with the paintbrush, in the direction from heel to toe. The paw withdrawal response was scored according to the following criteria: score = 0, walking away or occasionally very brief paw lifting (≤1 s); score = 1, sustained lifting (>2 s) of the stimulated paw toward the body; score = 2, a strong lateral lifting above the level of the body; score = 3, flinching or licking of the affected paw. Average scores for each mouse were obtained from three stimulations at intervals of at least 3 min.
Cold allodynia was measured by applying a drop of acetone to the midplantar area of the hindpaw and monitoring the behavioral responses for 1 min42,43. The cumulative time spent in pain behavior (paw licking, withdrawal, flinching, etc.) was recorded. The test was repeated three times at intervals of at least 15 min to obtain an average time in seconds for each animal.
Ongoing pain was tested using a conditioning paradigm44 with retigabine (R-100, Alomone Laboratory) as the conditioned stimulus to briefly relieve pain36,45,46. Mice were first allowed to freely explore the conditioned place preference apparatus, consisting of two chambers (one dark and one light) connected by a hallway (Stoelting), for 15 min. The time spent in the light chamber was recorded. During the conditioning phase, mice were first administered saline (i.p.) and kept in the dark chamber for 20 min. Then, 3 h later, the analgesic retigabine was administered (10 mg kg−1; i.p.) and, after 10 min, the mice were placed in the light chamber for 20 min. The conditioning was completed over four consecutive days. On the fifth day, the mice were again allowed to freely explore the apparatus for 15 min without any retigabine or saline injections. Data are presented as the difference in time spent in the light (retigabine-paired) chamber during the drug-free test on day 5 minus time spent in the light chamber at baseline (preconditioning phase). A mouse with spontaneous pain should show an increase in time spent in the light chamber that was paired with retigabine than it did in the preconditioning phase.
To examine numbness, we used a modified protocol of the adhesive removal test36,47. A round adhesive patch (3/16-inch Teeny Tough-Spots, USA Scientific) was placed on the plantar surface of the hind paws. The latency to attend to the patch (for example, shaking or attempted removal) was recorded within a 15-min testing time.
Statistics
All statistical results are reported in Supplementary Table 5. Differences between in vitro concentration–response relationships were determined by comparing the slopes of fitted linear functions for shared parameters using extra sum-of-squares F-tests. Linear models were selected as the pharmacologically relevant concentrations tested were within the linear range of the concentration–response functions. Differences in gene expression, protein levels and skin temperatures were analyzed by t-tests or one-way or two-way analysis of variance (ANOVA) followed by Dunnett’s or Tukey’s post hoc tests as appropriate. Von Frey and acetone data were analyzed by repeated-measures two-way or three-way ANOVA with Tukey or Šidák’s post hoc tests, one-way ANOVA with Dunnett’s post hoc tests or unpaired t-tests where appropriate. Brush data were analyzed by Friedman with Dunn’s post hoc tests, Kruskal–Wallis with Dunn’s post hoc tests or Mann–Whitney tests where appropriate. Conditioned place preference data were analyzed by two-way ANOVA with Tukey’s post hoc tests. Data from the adhesive removal test were analyzed by unpaired t-test. Analyses were performed using Prism version 10.2.2 (GraphPad). P < 0.05 was considered statistically significant. Parametric data are expressed as the mean ± s.d. and nonparametric data are presented as the median ± range.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.