Ethical statement
The research presented here complies with all relevant ethical regulations. All experiments involving animals were reviewed and approved by the Animal Care and Use Committee (ACUC) at the University of California, Berkeley before commencing the study. Housing, maintenance and experimentation of the mice were carried out with strict adherence to ethical regulations set forth by the ACUC at the University of California, Berkeley.
Plasmid construction
Plasmids used for the expression of different Cas proteins in this study were built on the basis of a pCold vector. The inserts encoding Cas proteins contain an N-terminal CL7 tag followed by an HRV-3C protease cleavage site and a C-terminal 6xHis-tag following another HRV-3C protease cleavage sequence. The insert for the final NLS-GeoCas9(R1W1)-2NLS protein contains an N-terminal sequence consisting of different tags, His6-CL7-MBP (maltose-binding protein) followed by an HRV-3C protease cleavage site. The cloning reactions were carried out in a 50-μl reaction containing 1 ng of template plasmid, 1.25 μl of 10 mM dNTP and 1.25 μl of 10 μM each primer using Phusion high-fidelity DNA polymerase (New England Biolabs (NEB), M0530L). After PCR, the reactions were treated with 1 μl of DpnI (NEB, R0176L) for 1 h at 37 °C before gel purification. The plasmids were ligated by Gibson assembly (NEB master mix, E2611L) of the plasmid backbone and insert sequences. The sequences of all the plasmid constructs were confirmed by full plasmid sequencing (Plasmidsaurus).
Nucleic acid preparation
All the DNA and RNA oligos used in this study, unless otherwise indicated, were purchased from IDT and passed the quality control standard set by IDT. The HM sgRNAs used in the study were laboratory-purified by PAGE. Some of the sgRNAs and ssDNA HDR templates purchased from IDT possess chemical modifications at the 3′ or 5′ ends (Supplementary Tables 2 and 3). The mRNA encoding 2NLS-iGeoCas9(C1)-2NLS was purchased from TriLink and purified with a silica membrane.
Lipid material preparation
Commercial lipid materials used in this study were purchased from BroadPharm, Avanti Polar Lipids and Cayman Chemical. Acid-degradable lipids, ADP, ADC, Pep-1k and Pep-2k, were laboratory-synthesized following the procedures in our previous publication47.
IVT of sgRNA
Four sgRNAs (UM-tdTom-g3(23), UM-tdTom-g7(23), UM-gPCSK9 and UM-gSFTPC) used in this study were prepared in milligram scale through IVT using HiScribe T7 high-yield RNA synthesis kit (NEB, E2040S). Following the general protocol provided by the supplier, each IVT reaction (1.2–1.4 ml) uses one RNA synthesis kit together with a dsDNA template (30–50 ug) encoding the sgRNA sequence under a T7 promoter. The IVT reaction mixture was incubated at 37 °C for 10–12 h, then treated with DNAse I (100 units; NEB, M0303S) and incubated for another 3–4 h before being quenched by 2× STOP solution containing formamide, bromophenol blue, xylene cyanol and EDTA. Urea–PAGE was used for sgRNA purification and the gel fraction containing the desired sgRNA was crushed into fine pieces and subjected to RNA extraction at 4 °C overnight using sodium acetate buffer (300 mM, pH 5.0). The extracted sgRNA was concentrated using an Amicon ultracentrifugal filter (10-kDa cutoff) to a total volume of 3–5 ml and the concentrated RNA solution was treated with 10 ml of cold isopropanol to allow the sgRNA to precipitate at −20 °C over 6 h. The sgRNA was pelleted by centrifuge and washed using cold 70% ethanol three times. The isolated sgRNA was further dissolved in 1× rCutsmart buffer (1 ml; NEB, B6004S), subjected to terminal triphosphate removal using calf intestinal alkaline phosphatase (100 units; NEB, M0525S) and incubated at 37 °C for 6 h. The reaction mixture was then diluted with sodium acetate buffer (4 ml, 300 mM, pH 5.0) and subjected to phenol–chloroform (5 ml, saturated, pH 5–6) extraction by vigorous vortexing and centrifugation; the aqueous phase was further washed with chloroform (5 ml) by vigorous vortexing and centrifugation three times. The sgRNA-containing aqueous solution was finally subjected to RNA precipitation and isolation; the pellet was dried in the open air to give purified sgRNA.
Purified IVT sgRNAs were dissolved in an endotoxin-free storage buffer (500 μl; 25 mM NaPi, 150 mM NaCl and 200 mM trehalose at pH 7.50). sgRNAs were reannealed by incubation at 64 °C for 5 min, followed by gradual cooling to room temperature. The sgRNA concentration was Nanodrop-determined (after 10–50× dilution). The final yields of GeoCas9 sgRNA by IVT were as follows: UM-tdTom-g3 and UM-tdTom-g7, 4–5 mg per reaction; UM-gPCSK9 and UM-gSFTPC, 8–12 mg per reaction.
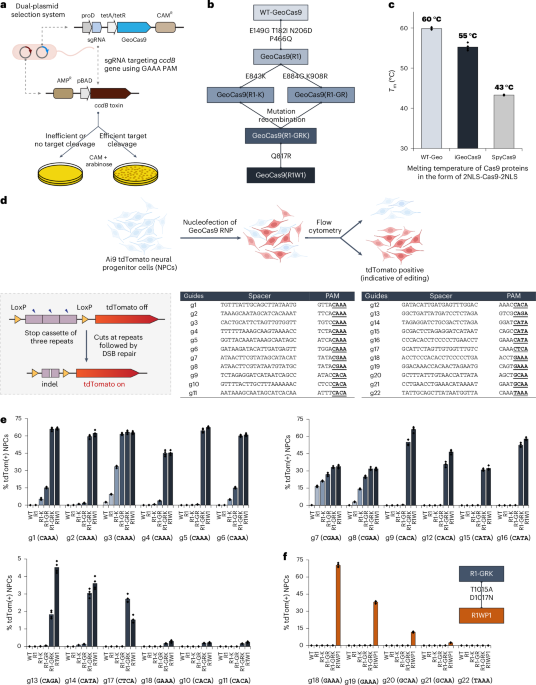
Directed evolution of GeoCas9
A chloramphenicol-resistant (CAM+) bacterial expression plasmid was built to have the insert gene of GeoCas9 together with its corresponding sgRNA that targets the ccdB gene in the selection plasmid with a PAM of GAAA (g6). Libraries of GeoCas9 mutants were generated by error-prone PCR to introduce random mutagenesis in three different regions (BH-Rec, RuvC-HNH-WED and WED-PI). The error-prone PCR (with an error rate of 3–5 nucleotide mutations per kilobase) was carried out with Taq DNA polymerase (NEB, M0273S) in a reaction containing 2 µl of 10 mM primers, 1.5 µl of 10 mM MnCl2 and 2 ng of template plasmid. The plasmid libraries were generated by ligating the mutated fragments with the remaining part of the plasmid through Gibson assembly. The plasmid libraries (~100 ng DNA after clean-up) were electroporated into 50 µl of electronically competent cells made from E. coli strain BW25141(DE3) that contained the selection plasmid encoding the arabinose-inducible ccdB toxin gene. After recovery of the electroporated bacteria in 750 µl of sulfur-oxidizing bacteria for 1.5 h at 30 °C, the bacteria culture was concentrated; 1% of the total culture was plated onto a Petri agar dish containing only CAM (as control) and the remaining culture was plated on another Petri agar dish containing both arabinose and CAM. Positive colonies that grew on the plates containing both arabinose and CAM were collected in a pool, retransformed (with ~2 ng of plasmid) and replated (100 µl of transformed culture on both control and selection plates). Plasmids of individual colonies from the replated plate were sequenced to obtain mutational information. Validation of the positive clones in the bacterial assay followed the same procedure.
Protein expression and purification
All the proteins in this study were expressed in E. coli BL21 (DE3) cells (Sigma-Aldrich) cultured in 2× YT medium supplemented with ampicillin. The cultivation was carried out at 37 °C with a shaking speed of 160 r.p.m. after inoculation with an overnight starter culture in LB medium containing ampicillin at a ratio of 1:40. When the optical density at 600 nm of the culture reached 0.8–0.9, the culture was cooled down to 4 °C on ice. The expression of Cas proteins was induced by the addition of IPTG to a final concentration of 0.1 mM and incubated at 15.8–16 °C with a shaking speed of 120 r.p.m. for 14–16 h.
To purify the Cas (or fusion) proteins, the cultured cells were harvested and resuspended in lysis buffer (50 mM Tris-HCl, 20 mM imidazole, 1.2 M NaCl, 10% (v/v) glycerol, 1 mM TCEP and cOmplete protease inhibitor cocktail tablets (Millipore Sigma, 1 tablet per 50 ml) at pH 7.5), disrupted by sonication and centrifuged at 35,000g for 45 min. Ni-NTA resin was treated with the supernatant at 4 °C for 60 min, washed with wash buffer 1 (lysis buffer without protease inhibitor cocktail tablet), and eluted with elution buffer (50 mM Tris-HCl, 300 mM imidazole, 1.2 M NaCl, 10% (v/v) glycerol and 1 mM TCEP at pH 7.5) to give crude His-tagged Cas proteins. The nickel elution was then subjected to Im7-6B resin in a slow gravity column repeatedly (3–4 times). The Im7-6B resin was washed with wash buffer 2 (50 mM Tris-HCl, 1.2 M NaCl, 10% (v/v) glycerol and 1 mM TCEP at pH 7.5) before being treated with HRV-3C protease (1% weight to crude Cas protein) for 2–2.5 h to release the Cas proteins from the CL7 and 6xHis-tags. Heparin affinity column was used to further purify the desired proteins. The protein fractions were collected, concentrated and stored in the storage buffer (25 mM NaPi, 150 mM NaCl and 200 mM trehalose at pH 7.50) after buffer exchange. The final yields of different Cas proteins (all with two copies of NLS at both N and C termini) were as follows: wild-type GeoCas9, ~10 mg per 1 L of culture; GeoCas9 mutants, in a range of 2–10 mg per 1 L of culture; SpyCas9, ~4 mg per 1 L of culture; iCas12a, ~30 mg per 1 L of culture; PE2 (nSpyCas9-RT), 1–2 mg per 1 L of culture.
The purification of NLS-GeoCas9(R1W1)-2NLS and 2NLS-GeoCas9(R1-GRK)-2NLS proteins was slightly different after Ni-NTA resin purification. The nickel elution was subjected to dialysis against dialysis buffer (50 mM Tris-HCl, 10 mM imidazole, 1.2 M NaCl, 10% (v/v) glycerol and 1 mM TCEP at pH 7.5) containing HRV-3C protease (1% weight to crude Cas protein) for 12–15 h. The tag-cleaved protein was then loaded to a heparin column and washed with 80 column volumes of buffer containing 0.1% Triton X-114 at 4 °C to minimize endotoxin impurities. The protein fractions were collected, concentrated and subjected to further purification using a size-exclusion column in an endotoxin-free manner. The purified protein was stored in an endotoxin-free storage buffer (25 mM NaPi, 150 mM NaCl and 200 mM trehalose at pH 7.50). The final yields of the desired GeoCas9 mutants were 3–5 mg per 1 L of culture.
Measurement of protein melting temperatures
Protein melting temperatures were measured using the thermal shift assay (GloMelt, 33021). The assay was performed on a qPCR system with a temperature increase rate of 2 °C min−1. The protein melting temperatures were determined as the peak values in the derivative curves of the melting curves.
Cell lines and culture conditions
NPCs were isolated from embryonic day 13.5 Ai9 tdTomato homozygous mouse brains. Cells were cultured as neurospheres at 37 °C with 5% CO2 in NPC medium containing DMEM/F12 (Gibco, 10565018) with GlutaMAX supplement, sodium pyruvate, 10 mM HEPES, nonessential amino acids (Gibco, 11140076), penicillin and streptomycin (Gibco, 10378016), 2-mercaptoethanol (Gibco, 21985023), B-27 without vitamin A (Gibco, 12587010), N2 supplement (Gibco, 17502048) and growth factors bFGF (BioLegand, 579606) and EGF (Gibco, PHG0311) (both 20 ng ml−1 as final concentration). NPCs were passaged using the MACS neural dissociation kit (Papain, 130-092-628) following the manufacturer’s protocol. bFGF and EGF were refreshed every 3 days and cells were passaged every 4‒5 days. Precoating with a coating solution containing poly(dl-ornithine) hydrobromide (Sigma-Aldrich, P8638), laminin (Sigma-Aldrich, 11243217001) and fibronectin bovine plasma (Sigma-Aldrich, F4759) was required for culturing cells in 96-well plates.
HEK293T and HEK293T EGFP cells were grown in a medium containing DMEM (Gibco, 10569010), high glucose, GlutaMAX supplement, sodium pyruvate, 10% FBS, penicillin and streptomycin (Gibco, 10378016) at 37 °C with 5% CO2. Cells were passaged every 3 days.
16HBEge cells were grown in a medium containing MEM (Gibco, 11090099), 10% FBS, penicillin and streptomycin (Gibco, 10378016) at 37 °C with 5% CO2. T75 flasks precoated with a coating solution containing LHC-8 basal medium (Gibco, 12677-027), 7.5% BSA (Gibco, 15260-037), bovine collagen solution, type 1 (Advanced BioMatrix, 5005) and fibronectin from human plasma (Thermo Fisher Scientific, 33016-015) were used for culturing 16HBEge cells. Cells were passaged every 4‒5 days. Precoating was required for culturing cells in 96-well plates.
RNP assembly
For cell culture experiments, RNPs were assembled at a 1.2:1 molar ratio of sgRNA (IDT or IVT) to Cas protein in a supplier-recommended buffer (for nucleofection) or a phosphate buffer (25 mM NaPi, 150 mM NaCl and 200 mM trehalose at pH 7.50) immediately before use; it is crucial to slowly add the Cas protein solution to the sgRNA solution while swirling (the reverse addition order can cause RNP aggregate formation). The solution was incubated for 15–25 min at room temperature or 5–10 min at 37 °C. For nucleofection, RNPs were further complexed with Alt-R Cas9 electroporation enhancer (100-nt ssDNA; IDT, 10007805) with a 1:1 molar ratio of enhancer to RNP in supplier-recommended buffers (Lonza). For LNP assembly, RNPs ± ssDNA were further diluted with a neutral solution of PBS and water (1:1, pH 7.3‒7.5) or an acidic solution of sodium citrate (10 mM, pH 5.0) to a certain RNP concentration.
Genome editing with different cell lines
For nucleofection, 250,000 NPCs or 200,000 HEK293T cells were nucleofected with 100 pmol (or other doses if specified) of preassembled RNP (with 100 pmol of enhDNA) with program codes of EH-100 and CM-130, respectively, according to the manufacturer’s instructions. Lonza SF (for HEK293T cells) and P3 (for tdTomato NPCs) buffers were used for the preparation of nucleofection mixtures (with a total volume of 20 µl). Then, 10% of the nucleofected cells were transferred to 96-well plates. The culture medium for NPCs was refreshed after 3 days; HEK293T cells were split with a ratio of 5:1 after 3 days. Cells were harvested for analysis after further incubation at 37 °C for 2 days.
For LNP delivery, 4,000–6,500 cells per well were seeded in 96-well plates 48 h before LNP treatment (HEK293 cells, 4,000–5,000; NPCs, 5,000–6,000; 16HBEge cells, 6,000–6,500). The culture medium was refreshed 24 h after LNP treatment. HEK293T cells were split after two additional days with a ratio of 1:1 to 2:1 based on cell confluency. Cells were harvested for analysis after a total incubation time of 4–5 days (upon signal maturation for tdTom NPCs and HEK293 EGFP cells).
Cell viability was determined on the basis of the counts of live cells (stained with trypan blue) at certain times after treatment with LNPs in comparison to cells treated with PBS (negative control).
Flow cytometry
Cell fluorescence was assayed on an Attune NxT acoustic focusing cytometer (Thermo Fisher Scientific) equipped with a 554-nm excitation laser and 585/16 emission filter (tdTomato), 488-nm excitation laser and 530/30 emission filter (EGFP) and 400-nm excitation laser and 440/50 emission filter (BFP) and corresponding setup for cell type analysis based on the antibody fluorophores. Flow data were analyzed using Attune Cytometric Software (version 5.1.1), FlowJo (version 10.7.1), Excel (version 2408) and Prism 9 (version 9.4.1).
LNP assembly and delivery experiment setup
LNP solutions with a total volume of less than 200 μl were prepared by pipet mixing; LNP solutions with higher volumes were prepared using a microfluidic mixing device, NanoGenerator Flex-M (PreciGenome).
For the preparation of standard and cationic LNPs at small scales, RNPs were assembled by mixing iGeoCas9 and sgRNAs at a molar ratio of 1:1.2 and incubated for 20‒30 min at room temperature. For HDR experiments, the assembled RNPs were further mixed with ssDNA templates at a molar ratio of 1:1 (ssDNA to RNP). The RNP stock solutions were diluted with an aqueous solution (PBS and water, 1:1, with 5 mM DTT, pH 7.3‒7.5) to give a final RNP concentration of 5.0 or 7.5 μM. The lipid stock solutions in ethanol and DMSO were prepared at a total lipid concentration of 10‒12 mg ml−1. LNPs were assembled by pipet mixing with a volume ratio of 4:1 (aqueous to organic) and incubated at room temperature for 1 h before being diluted with PBS (3× volume of the LNP solution) to give an LNP stock solution with RNP concentrations of 1.0 or 1.5 μM. For cell culture experiments, the LNP solutions were diluted with the corresponding culture medium (9× volume of the LNP solution in PBS) and then used to treat cultured cells (in 1:1 volume ratio) with a final RNP concentration of 50 or 75 nM RNP (for example, 5.0 or 7.5 pmol of RNP in 100 μl of culture medium).
For long-term storage of standard and cationic LNPs at 4 °C, DTT was excluded during LNP assembly. A solution of DTT (5 mM) in PBS was used to activate LNPs right before the in vitro delivery experiments.
For the preparation of FX12 and FC8 LNPs at small scales, RNPs were assembled by mixing iGeoCas9 and sgRNAs at a molar ratio of 1:1.2, incubated for 10 min at 37 °C, then complexed with 100-nt enhDNA (1.5 equivalents to RNP), and incubated for another 10 min at 37 °C, giving a stock solution of RNP with a concentration of 12‒15 μM in the storage phosphate–trehalose buffer. The RNP stock solution was then diluted with sodium citrate buffer (10 mM, pH 5.0) to give a final RNP concentration of 1.06 μM (final pH of ~5.2). The lipid stock solutions in ethanol and DMSO were prepared at a total lipid concentration of 10 mg ml−1. LNPs were assembled by pipet mixing with a volume ratio of 4:1 (aqueous to organic) and incubated at room temperature for 20‒30 min before being neutralized and diluted by PBS (3.24× volume of the LNP solution) to give an LNP stock solution with an RNP concentration of 0.2 μM. The LNP stock solution could be further diluted to give certain doses used for in vitro RNP delivery experiments. FC8 LNPs were DTT-activated during PBS dilution. For cell culture experiments, the LNP solutions were diluted with the corresponding culture medium (9× volume of the LNP solution in PBS) and then used to treat cultured cells. For instance, 5 μl of the LNP stock solution (with an RNP concentration of 0.2 μM) was diluted with 45 μl of culture medium and used to treat mammalian cells in 50 μl of culture medium in a 96-well plate, giving a final RNP concentration of 10 nM (1.0 pmol of RNP in 100 μl of culture medium).
For the microfluidic preparation of LNPs, RNPs were assembled by mixing iGeoCas9 and sgRNAs with a molar ratio of 1:1.2, incubated for 10 min at 37 °C, then complexed with 100-nt enhDNA (1.5 equivalents to RNP), and then incubated for another 10 min at 37 °C, giving a stock solution of RNP with a concentration of 12‒15 μM in the storage phosphate–trehalose buffer. The RNP stock solution was then diluted with sodium citrate buffer (10 mM, pH 5.0) to give a final RNP concentration of 1.25 μM (final pH of ~5.2) or 0.625 μM (final pH of ~5.1). The lipid stock solutions in ethanol and DMSO were prepared with a total lipid concentration of 10 mg ml−1 (for FX12m formulation) and 5 mg ml−1 (for FX8m formulation). FX12m LNPs were microfluidic-assembled with a volume ratio of 4:1 (aqueous to organic) at a flow rate of 3 ml min−1; FC8m LNPs were microfluidic-assembled with a volume ratio of 4:1 (aqueous to organic) at a flow rate of 2 ml min−1. The assembled LNPs were incubated at room temperature for 20‒30 min before being subjected to dialysis against PBS using a dialysis membrane with an MW cutoff of 10 kDa (Thermo Fisher Scientific) overnight at 4 °C. Upon dialysis, the LNPs were concentrated by ultrafiltration using Amicon ultracentrifugal filter with an MW cutoff of 100 kDa (Millipore). FC8m LNPs were DTT-activated during the concentration step. The filter was washed three times with PBS to collect the remaining LNPs absorbed on the filter membrane. The combined LNP solution was diluted with PBS to a certain volume used for animal experiments.
Dynamic light scattering (DLS) assay
The size distribution of RNP particles or LNPs was measured using Zetasizer (version 7.13, Malvern Panalytical; He–Ne Laser, λ = 632 nm; detection angle = 173°).
RNP encapsulation rate measurement
Quant-iT RiboGreen RNA reagent (R11491) was used to estimate RNP encapsulation efficiency. LNP and lysed LNP (using 1% Triton X-100) samples were diluted using TE buffer to an estimated total nucleic acid concentration of 0.5‒2.0 ng μl−1. The diluted (lysed) LNP samples were mixed with the RiboGreen reagent (1:1,000 dilution in TE buffer) in a volume ratio of 1:1 (100 μl + 100 μl) and incubated at room temperature in the dark for 2 min before fluorescent signal measurement with the emission wavelength of 500/525 nm. The unencapsulated RNP proportion was estimated as the ratio of the signal intensity (with blank signal subtracted) of intact LNP to lysed LNP samples, thus giving the corresponding RNP encapsulation rate.
Cryo-transmission electron microscopy (cryo-TEM) image acquisition and processing
For cryo-TEM imaging, 3 µl of LNP suspension was added to a glow-discharged R2/2 Quantifoil Cu Grid (Ted Pella). Samples were incubated for 20 s and blotted for 4 s (blotting force = 5) in a 4 °C high-humidity chamber. After incubation, the samples were immediately plunge-frozen using an FEI Mark IV Vitrobot (FEI), resulting in vitreous ice. The samples were then imaged with an FEI Talos Arctica at 200 kV under low-dose conditions using a bottom-mounted K3 camera (Gatan) at ×36,000 magnification (0.5705 Å per pixel). Images were analyzed with Cryo-SPARC software (version 4.5.3) and representative images were selected from multiple viewfields and grids.
In vivo genome editing
Retro-orbital injections of LNPs consisting of different lipid formulations were performed with Ai9 tdTomato mice (C56BL/6J, Jackson Laboratory) and wild-type mice (BALB/c, Jackson Laboratory), 10–16 weeks old, weighing 18–20 g (male or female). The mice were killed and all tissues were collected for further analysis 2 weeks (Ai9 mice) or 10 days (wild-type mice) after LNP injection.
For flow analysis, isolated tissues were minced using a sterile blade and then subjected to digestion with collagenase type I (0.1 mg ml−1 as the final concentration; Gibco, 17018029) in 1 ml of Hanks’ balanced salt solution buffer (Gibco, 14175095) supplemented with 5 mM Ca2+ at 37 °C for 2 h with gentle shaking. Next, the digested solution was filtered using a 70-μm filter and quenched with PBS containing 2% FBS. A cell pellet was obtained by centrifuging for 5 min at a speed of 1,500g at 4 °C. The supernatant was removed and the cell pellet was resuspended in 1 ml of PBS containing 2% FBS, which could be used for flow analysis.
For cell type analysis, the dissociated tissue cells (100 μl) were incubated with corresponding antibodies (1:200 dilution) for 1 h in the dark at 4 °C. The stained cells were washed three times with 500 μl of PBS and then resuspended in 500 μl of PBS for flow cytometry analysis. The antibodies used for liver cell types were Alexa Fluor 647 anti-mouse CD95 (Fas) (BioLegend, 152620, for hepatocytes), Alexa Fluor 647 anti-mouse F4/80 (BioLegend, 157314, for macrophages) and Alexa Fluor 488 anti-mouse CD31 (BioLegend, 102414, for endothelial cells); the antibodies used for lung cell types were Alexa Fluor 647 anti-mouse CD326 (Ep-CAM) (BioLegend, 118212, for epithelial cells), Alexa Fluor 488 anti-mouse CD31 (BioLegend, 102414, for endothelial cells) and Pacific blue anti-mouse CD45 (BioLegend, 157212, for immune cells).
For analysis by imaging, tissue blocks were embedded into optimal cutting temperature compounds (Sakura Finetek) and cosectioned (8 μm) on a Cryostat instrument (Leica Biosystems) to prepare tissue sections. The mounted tissue slices were stained with DAPI before microscopy imaging. Images of tissue slices were taken using the Leica DMi8 microscope and analyzed using the Leica Application Suite X program (version 3.9.1.28433).
For analysis by NGS or DNA gel assays, dissociated tissue cells were collected and treated with Quick Extraction solution (Epicentre) to lyse the cells (65 °C for 20 min and then 95 °C for 20 min). The cell lysates were directly used for gene amplicon prep by PCR.
NGS
Edited cells were harvested and treated with Quick Extraction solution (Epicentre) to lyse the cells (65 °C for 20 min and then 95 °C for 20 min). The cell lysates were directly used for gene amplicon prep by PCR. Amplicons of genomic targets were PCR-amplified in the presence of corresponding primers, which were designed to have no overlap with their corresponding donor ssDNA sequence in the case of HDR. The PCR products were purified with magnetic beads (Berkeley Sequencing Core Facility) before being subjected to NGS with MiSeq (Illumina) at 2 × 300 bp with a depth of at least 20,000 reads per sample. The sequencing reads were subjected to CRISPResso2 (https://github.com/pinellolab/CRISPResso2) to quantify the levels of indels and HDR. Subsequent data analysis and presentation were performed with Excel (version 2408) and Prism 9 (version 9.4.1).
Immunogenicity assessment
Wild-type BALB/c mice (male or female), 10–16 weeks old, weighing 18–20 g, were used for cytokine measurement experiments. For LNP complexes based on FX12m or FC8m formulations with RNP cargo or as empty vectors, RNP-only solution, PBS (negative control) and LPS (lipopolysaccharide, with a dose of 1 mg kg−1; positive control) were administered by the retro-orbital route (intravenous). LNPs (with or without RNPs) were injected at doses following the experimental setup (for example, 4.6 mg kg−1 RNP for FX12m formulation). Injections were performed with a total volume of 150 μl per mouse.
At time points of 6 and 24 h, the first two batches of mice were killed and corresponding blood samples were collected in heparin and centrifuged at 1,500g for 10 min at 4 °C. The levels of cytokines, including interleukin 2 (ELISA kit from R&D Systems, DY402-05), interleukin 6 (ELISA kit from R&D Systems, DY406-05), macrophage inflammatory protein 2 (ELISA kit from R&D Systems, DY452-05) and tumor necrosis factor (ELISA kit from R&D Systems, DY410-05), in the plasma were determined on the basis of ELISA assays following the manufacturer’s protocols (R&D Systems).
Another batch of mice were killed 2 weeks after injection and blood samples were collected in heparin and centrifuged at 1,500g for 10 min at 4 °C. The levels of liver damage enzymes, including alanine aminotransferase (ELISA kit from Abcam, ab282882), aspartate aminotransferase (ELISA kit from Abcam, ab263882) and transglutaminase 2 (ELISA kit from RayBiotech, ELM-TGM2-1), in the plasma were determined on the basis of ELISA assays following manufacturers’ protocols (Abcam or RayBiotech).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.