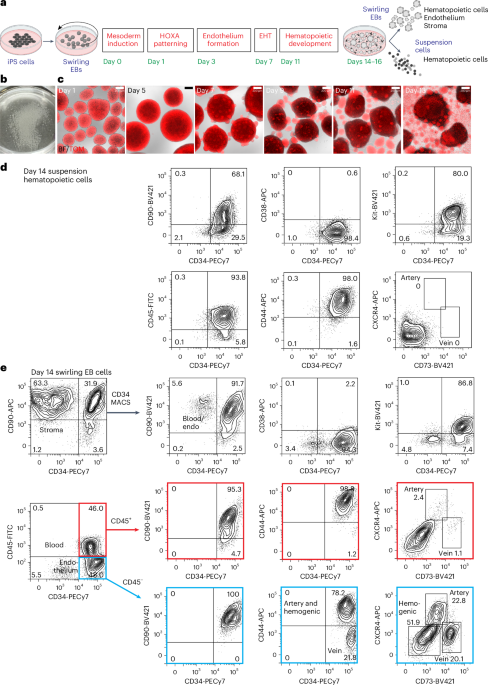
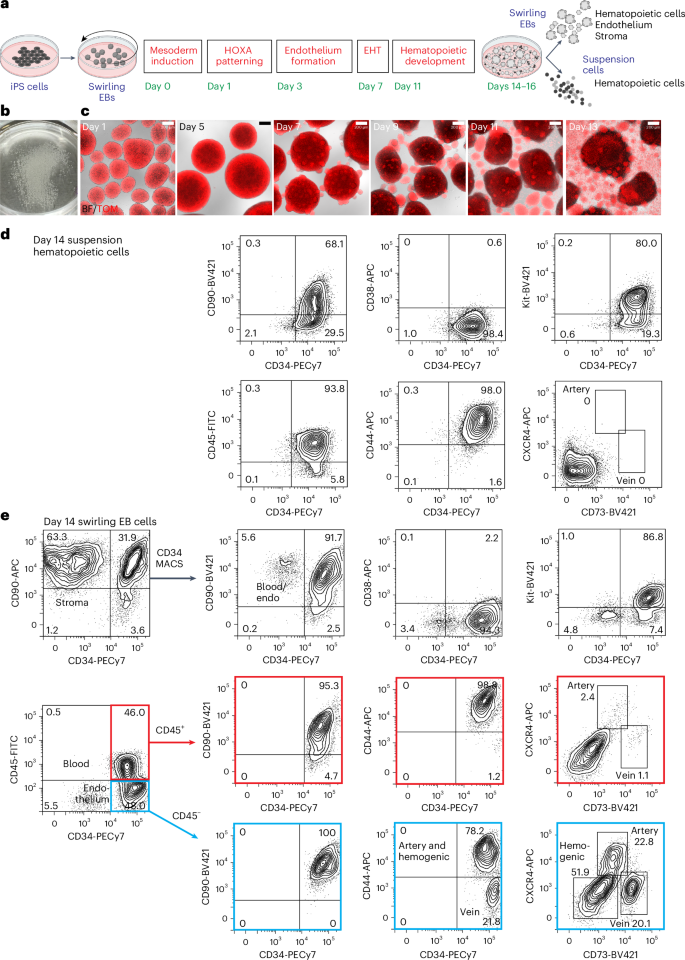
Differentiation of iPS cells to CD34-expressing hematopoietic cells
For all differentiation protocols, iPS cells were dissociated and seeded into dishes that were incubated on a rotating platform, allowing the formation of swirling embryoid bodies (EBs) that differentiated to hematopoietic cells13,22 (Fig. 1a,b and Supplementary Results 1; see Extended Data Fig. 1a for an overview of differentiation protocols and Methods for details of growth factor combinations). Mesoderm was induced for 24 h, patterned for 2 days to induce the expression of HOXA genes18 and differentiated to hemogenic endothelium from days 3 to 7. Cells undergoing an endothelial-to-hematopoietic transition protruded from the surface of the EBs, reminiscent of intra-arterial hematopoietic clusters of blood cells emerging from the embryonic aorta10,11 (Fig. 1c). These cellular accumulations broke away from the EBs, shedding blood cells into the medium from day 11 (Fig. 1c). Cultures on day 14 comprised a dominant blood cell suspension with most cells expressing CD34, CD90, CD44 and Kit (Fig. 1d and Supplementary Fig. 1). The EB-derived fraction consisted of the stroma, the endothelium and hematopoietic cells that had not yet shed into the medium (Fig. 1e and Supplementary Fig. 1). A small proportion of the hematopoietic cells expressed CXCR4 or CD73, reflecting their recent emergence from an endothelial precursor (Fig. 1e). From days 14 to 16, the suspension hematopoietic cells were harvested and cryopreserved (Fig. 1d). In some experiments, CD34+ cells enriched from EBs by magnetic-activated cell sorting (MACS) (Fig. 1e) were also cryopreserved.
a, Swirling EB differentiation protocol indicating differentiation stages transitioning from undifferentiated iPS cells to hematopoietic, endothelial and stromal cells. Growth factors for each stage are shown in Extended Data Fig. 1a and the Methods. EHT, endothelial-to-hematopoietic transition. Partially created using BioRender.com. b, A 60-mm dish on day 7 showing hundreds of swirling EBs. c, Overlaid bright-field (BF) and tandem TOMATO (TOM) fluorescence images of developing swirling EB cultures. Scale bar, 200 µm. d, Flow cytometry of day 14 suspension hematopoietic cells showing the expression of surface CD45, CD34, Kit, CD44 and CD90. e, Dissociated day 14 swirling EB cells were typically enriched to >90% CD34+ endothelium and blood using MACS. These cells comprised CD45+ blood cells (profiles with red borders) and CD45− endothelium (profiles with blue borders). The endothelium was categorized as arterial, venous or hemogenic on the basis of the expression of CD34, CD44, CXCR4 and CD73 (ref. 15). The flow cytometry results in d,e are from one representative experiment of more than 20 experiments performed.
MLE cells require retinoids during iPS cell differentiation
We screened combinations of CHIR, Activin A, bone morphogenetic protein 4 (BMP4) and a retinoid during the mesoderm induction and patterning stages (screening protocol 1; Methods and Extended Data Fig. 1a,b) to determine whether any supported the generation of engraftable human hematopoietic cells. CD34+ hematopoietic cells were generated from an iPS cell line constitutively expressing a tandem TOMATO fluorescent protein (RM TOM) (Fig. 1c)23 and cryopreserved before thawing and injection into the tail vein of NOD,B6.Prkdcscid Il2rgtm1Wjl/SzJ KitW41/W41 (NBSGW) mice24, mimicking the workflow of clinical HSC transplantation (Fig. 2a,b). In this series of experiments, groups of mice (totaling 134, denoted cohort 1) were injected with cells differentiated under one of 12 mesoderm induction and patterning protocols in screening protocol 1 (Supplementary Results 2, Fig. 2a–f, Extended Data Fig. 1b–d and Supplementary Tables 1–3 and 13). Some mice (12/134) were engrafted by stem cells displaying multilineage differentiation resulting in erythroid, myeloid and lymphoid reconstitution (denoted MLE). We found that most mice in which MLE occurred received cells in which the mesoderm was induced with 4 µM CHIR on day 0 and a pulse of a retinoic acid precursor (retinol (ROL) or retinyl acetate (RETA)) was included from days 3 to 5 of differentiation (Fig. 2a–e). Indeed, 17.6% (9/51) of mice transplanted with cells treated with the combination of 4 µM CHIR and retinoid showed MLE (Fig. 2f). There were over 80% human cells occupying the bone marrow in some of these MLE cohort 1 recipients (average: 46.5% ± 10.0% human cells in bone marrow and 11.9% ± 5.1% in spleen) (Fig. 2f and Supplementary Tables 1 and 13), highlighting the capacity for differentiation of the engrafting cells. All MLE mice in cohort 1 and subsequent transplant cohorts were engrafted with ≥0.1% human cells (Supplementary Tables 13 and 19). Hereafter, we refer to these functionally defined iPS cell-derived multipotent hematopoietic cells with the capacity to engraft multiple lineages over a long term as ‘iHSCs’.
a, Swirling EB differentiation protocol (screening protocol 1; Extended Data Fig. 1a) indicating mesoderm induction factors provided during the first day of differentiation and retinoids during endothelium formation from days 3 to 5 to generate the 12 differentiation conditions transplanted into mice in cohort 1. Numbers indicate the concentration of CHIR (CH) in µM and concentrations of BMP4 (B) and Activin A (A) in ng ml−1. b, Transplantation workflow showing the cryopreservation of CD34+ hematopoietic cells from the cell suspension along with MACS-isolated CD34+ cells from the EB. MACS-enriched EB cells were not collected for all experiments. Cryopreserved cells were thawed and transplanted immediately into NBSGW immune-deficient mice by tail-vein injection. Peripheral blood was analyzed at 12 weeks to screen for engraftment and hematopoietic tissues were analyzed for human cells at time points up to 24 weeks (Supplementary Tables 2 and 3). c, Scatter dot plot correlating the percentage of bone marrow (BM) human cells with differentiation conditions in cohort 1. Error bars, mean ± s.e.m. The number of mice receiving cells subjected to each mesoderm induction (n) is shown. The number of unengrafted (NEG) mice is indicated for each condition. d, Scatter dot plot correlating the concentration of CHIR during mesoderm induction with the phenotype of engrafted human cells in the BM (colored circles). The number of mice displaying an MLE phenotype differed between those receiving cells treated with 4CH and 1CH. *P = 0.03, determined by a two-sided Fisher’s exact test. Error bars, mean ± s.e.m. Data from the 4CH 3B5A and 4CH 30A mesoderm inductions were pooled. e, Scatter dot plot correlating the inclusion of retinoid (ROL or RETA) during iPS cell differentiation with the phenotype of engrafted human cells in the BM (colored circles). The number of mice displaying an MLE phenotype differed between those receiving cells treated with or without retinoid. ROL or RETA versus NIL (no retinoid). *P = 0.03, determined by a two-sided Fisher’s exact test. Error bars, mean ± s.e.m. Data from the 4CH 3B5A and 4CH 30A mesoderm inductions were pooled. f, Phenotypes in 42/51 mice transplanted with cells treated with the combination of 4 µM CHIR and retinoid (RET) that showed engraftment. In total, 9/51 (17.6%) transplanted mice showed MLE. Error bars, mean ± s.e.m.
Transcriptional similarity of in vitro differentiated iPS cells and human AGM
To search for transcriptional signatures accounting for functional differences between cells differentiated with and without retinoid and to allow comparisons to published datasets of human AGM, we performed single-cell RNA sequencing (scRNA seq) of differentiated iPS cells13,25. Two iPS cell lines were profiled, the RM TOM line described above and an independent line in which the mTagBFP2 fluorescent protein23 was expressed from the GAPDH locus of PB1.1 iPS cells26 (denoted PB1.1 BFP). Mesoderm was induced with 4 µM CHIR and 3 ng ml−1 BMP4 and 5 ng ml−1 Activin A (4CH 3B5A) or 30 ng ml−1 Activin A (4CH 30A), because our transplant results in cohort 1 identified that these conditions supported the generation of MLE mice (Fig. 2c). Because embryo data indicate that AGM-derived HSCs develop in a retinoid-replete milieu20, cultures were treated with RETA from days 3 to 5 (as was the case in cohort 1) or for a more prolonged period where RETA was added every 2 days from days 3 to 11, 13 or 14) (Fig. 3a). Control cultures were differentiated without added RETA. After 14 days, blood cells in suspension and disaggregated EBs were subjected to scRNA seq using the 10X Genomics platform. In total, 252,607 cells, comprising 12 RM TOM and 16 PB1.1 BFP samples, were analyzed. Uniform manifold approximation projection (UMAP) plots of integrated samples from both cell lines, followed by cluster analysis, allowed the allocation of cells to stromal, endothelial, hemogenic and hematopoietic lineages (Fig. 3b–e and Supplementary Table 4). The diverse lineages identified in scRNA seq indicate the considerable heterogeneity already present in the cultures, which was not obvious from the flow cytometric phenotype.
a, Swirling EB differentiation protocol showing the mesoderm induction and retinoid combinations used to differentiate RM TOM and PB1.1 BFP iPS cells. Each cell line was subjected to two mesoderm induction conditions, with 4 µM CHIR, 3 ng ml−1 BMP4 and 5 ng ml−1 Activin A (4CH 3BA5) or 4 µM CHIR and 30 ng ml−1 Activin A (4CH 30A), and three or four RETA exposure patterns. Samples were isolated from swirling EB and suspension hematopoietic cell fractions on day 14 of differentiation, leading to 28 samples subjected to scRNA seq. Partially created using BioRender.com. b,c, UMAP of integrated samples for individual lines (b) and following pooling of samples (c), showing the annotation of cell clusters allocated on the basis of cluster-specific gene expression (Supplementary Table 4). d,e, Feature plots depicting selected genes identifying tissue types (d) and hematopoietic cell lineages (e) in integrated samples. f, Feature plots depicting the expression of six human HSC signature genes13 in arterial (Art1), endothelial or stromal (En/Str), hemogenic (HE) and HLF+SPINK2+ cells from HSPC clusters 1–3 in integrated samples. The cell numbers and composition of clusters are provided in Supplementary Table 5. g, Violin plots showing the expression of selected stem cell genes in HLF+SPINK2+ cells from the HSPC cluster in CS14 and 15 embryos and from HLF+SPINK2+ cells from HSPC cluster 1 (c) in PB1.1 BFP and RM TOM cells. Cell numbers: CS14, 51; CS15, 70; PB1.1 BFP, 2,983; RM TOM, 1,112 (Supplementary Table 5). h, Comparison of the expression profiles of HLF+SPINK2+ cells from HSPC clusters from PB1.1 BFP and RM TOM cells to reference data from human embryonic-derived and CB-derived endothelial and hematopoietic cell populations, using the ACTINN machine learning algorithm to determine the percentage of iPS cell-derived hematopoietic cells displaying the greatest similarity to each reference dataset. Data stratified by retinoid treatment are shown for each cell line. The bar height represents the percentage of HLF+SPINK2+ putative iHSCs that map most closely to each reference sample. Cell numbers mapping to each reference sample are shown in Supplementary Table 10. EC, endothelial cell; VE, venous endothelium; AE, arterial endothelium; preHE, prehemogenic endothelium (representing aortic endothelium); HE, hemogenic endothelium; W, week; Plac, placenta; Ery, erythroid; Prog, progenitor; Meg, megakaryocyte; Mast, mast cell; Mono, monocyte; Mac, macrophage; Gran, granulocyte.
Reclustering cells within arterial, hemogenic and HLF+SPINK2+ cells within the HSC clusters confirmed that the HSC signature genes (RUNX1, MECOM, MLLT3, HLF, HOXA9 and SPINK2) recently identified in the human AGM13 (Supplementary Fig. 2a,b) were also expressed in iPS cell-derived cells (Fig. 3f and Supplementary Table 5). The percentage of cells expressing HSC signature genes and the level of expression of these genes were very similar in the RM TOM and PB1.1 BFP cell lines under both mesoderm induction conditions (4CH 3B5A and 4CH 30A) (Extended Data Fig. 2a). We also confirmed the expected pattern of HOXA gene expression in response to the SB and CHIR patterning in both cell lines (Extended Data Fig. 2b).
The addition of retinoids minimally impacted stem cell gene expression (Extended Data Fig. 2c) but notably influenced genes associated with retinoic acid metabolism such as CYP26B1, DHRS3, CRABP2, RARB and RARG, modulators of Wnt and fibroblast growth factor (FGF) signaling such as SHISA3, DKK1, RSPO1 and WNT4, as well as genes associated with vascular and hematopoietic development such as FOXC2 and CD38 (Supplementary Results 3, Supplementary Fig. 3 and Supplementary Tables 6–9). Many retinoid-responsive genes were only induced if the retinoids were included until at least day 11 of differentiation (Extended Data Fig. 2d).
We compared the transcriptional profiles of the iPS cell-derived HLF+SPINK2+ cells to similar HLF+SPINK2+ stem cell-like populations from human embryos at CS14 and CS15, examining the expression of a selected range of relevant genes (Fig. 3g). For a more extensive comparison between in vitro and human embryo-derived samples, we made use of the suite of scorecards developed in profiling studies of hematopoietic development in human embryos13 (Supplementary Results 4 and Extended Data Figs. 3 and 4). These studies benchmarked our iPS cell HLF+SPINK2+ cells against HLF+SPINK2+ CS14 and CS15 human embryo cells, demonstrating a high level of concordance between the transcriptional profiles across the nine scorecards of genes examined.
Lastly, we used a machine learning algorithm, ACTINN27, to compare the expression profiles of day 14 differentiated iPS cells to a human reference dataset comprising hematovascular cells from gestational day 22 to 24 (CS10–11) embryo and YS, day 29–36 (CS14–15) AGM, YS, embryonic liver and placenta and week 6, 8, 11 and 15 embryonic and fetal liver hematopoietic stem and progenitor cells (HSPCs) and cord blood (CB) stem and progenitor cells13. This analysis confirmed that HLF+SPINK2+ cells were most closely related to cells categorized as HSPCs in CS14–15 AGM, placenta and YS (Fig. 3h and Supplementary Table 10). Dissecting the allocation of cells to these categories from the two cell lines and the different durations of retinoid treatment revealed that cells derived from the RM TOM line mapped predominantly to the CS14–15 AGM HSPCs, whilst the PB1.1 BFP cells were more similar to CS14 YS and placental HSPCs. We can only speculate whether the greater overall proportion of RM TOM cells mapping to the CS14–15 AGM HSC sample was of functional importance. For both lines, the longer duration of retinoid increased the proportion of CS14–15 AGM HSPCs and decreased the CS14 YS and placental HSPCs (Fig. 3h).
MLE cells are generated from cultures treated with retinoids throughout differentiation
The ACTINN analysis identified the association of a longer duration of RETA treatment with a greater proportion of HLF+SPINK2+ iPS cell-derived cells that mapped to CS14–15 AGM HSPCs. We explored the functional ramifications of this observation by varying the duration of retinoid exposure in a second series of transplantation experiments. In ten experiments using cells sourced from six independent differentiations, 103 animals (cohort 2) were injected with differentiated RM TOM hematopoietic cells exposed to increasing durations of retinoid treatment (screening protocol 2 in Extended Data Fig. 1a, Fig. 4a,b and Supplementary Tables 2, 3 and 11). Data from mice receiving cells in which mesoderm was induced with 4CH 3B5A or 4CH 30A were pooled, given that both variations displayed a similar expression of HSC signature genes (Extended Data Fig. 2a). MLE was seen in 6/25 (24%) mice transplanted with cells treated with RETA from days 3 to 5, the duration of RETA that was successful for engraftment in cohort 1, and in 7/19 (36.8%) mice receiving cells exposed to RETA treatment from days 3 to 13, although the difference did not achieve statistical significance. We did not see MLE in mice receiving cells treated with RETA from days 3 to 7 or 9 and only in one of eight mice receiving RETA from days 3 to 15 of differentiation (Fig. 4b). While this suggests that retinoid signaling in hematopoiesis is temporally tightly regulated, we are cautious about overinterpreting these results because they were based on small numbers of animals transplanted with cells derived from only two differentiation experiments (Supplementary Table 11). All mice with MLE were analyzed >16 weeks after transplantation, with one exception (analyzed at 15.7 weeks). Notably, these experiments confirmed that prolonged treatment with RETA was compatible with the in vitro generation of MLE iHSCs from iPS cells, consistent with our transcriptomic data showing that prolonged exposure to a retinoic acid precursor was required for the expression of retinoid-responsive genes (Extended Data Fig. 2d) and embryo data indicating that AGM-derived HSCs develop in a retinoid-conditioned milieu13,20 (Supplementary Fig. 2c).
a, Swirling EB differentiation protocol (screening protocol 2; Extended Data Fig. 1a) showing the mesoderm induction and retinoid combinations used to differentiate RM TOM iPS cells for cohort 2 transplants. Cells were subjected to two mesoderm induction conditions and six retinoid exposure patterns before harvesting and cryopreservation on days 14–16. Partially created using BioRender.com. b, Scatter dot plot correlating human cells in the BM with the interval of retinoid (R) treatment during differentiation (shown as days) in cohort 2. Each circle represents one animal, color-coded to represent myeloid (M), myelo-lymphoid (ML), erythro-myeloid (EM) and erythro-myelo-lymphoid (MLE) patterns of engraftment. The number of mice receiving each duration of RETA (n) is shown. The number of unengrafted (NEG) mice is indicated. Data from 4CH 3B5A and 4CH 30A mesoderm inductions were pooled because they functioned similarly in the cohort 1 transplant experiments (Fig. 2c). Error bars, mean ± s.e.m. c, Confocal images of BM cells from an engrafted (m536) and unengrafted (m534) recipient. Scale bar, 50 µm. d–g, Flow cytometry profiles from BM (d), peripheral blood (PB; e), spleen (SPL; f) and thymus (THY; g) of a multilineage repopulated recipient (m490). d, Erythroid cells (CD43+GYPA+) were enriched in the TOM low (lo) BM fraction. The TOM high (hi) BM cells comprised CD19+ B cells, CD33+/CD13+ myeloid cells and CD45+CD34+CD38lo/− HSC-like cells (boxed in red). f, The SPL contained CD45+CD19+sIGM+ B cells. g, The THY contained immature CD45+CD3− thymocytes including CD4−CD8− cells, transitioning through immature CD4+ to CD4+CD8+ double-positive cell states, whilst CD45+CD3+ thymocytes included CD4+CD8+ double-positive and CD4+ and CD8+ single-positive cells.
We performed a similar series of experiments using the second transcriptionally profiled human iPS cell line, PB1.1 BFP, transplanting 79 mice in eight experiments derived from six independent differentiation experiments (cohort 3). Bone marrow engraftment was observed in 44.3% of recipients with predominantly myeloid-restricted engraftment, although one mouse demonstrated MLE after 19 weeks (Extended Data Fig. 5a–c and Supplementary Table 12). These data demonstrated that our differentiation protocol enabled the generation of MLE cells from a second independent iPS cell line, although the lower frequency of engraftment highlighted the requirement for protocol improvement.
MLE recipients of iHSCs showed MLE of hematopoietic tissues and establishment of a bone marrow stem cell compartment
We examined the contribution and lineage distribution of human cells in the bone marrow, spleen, thymus and peripheral blood of MLE animals identified in cohorts 1–3 in more detail (Fig. 4c–g and Extended Data Fig. 6). Human cells were present in the peripheral blood at 12 weeks after transplantation in MLE recipients (Extended Data Fig. 6a and Supplementary Table 13). Confocal analysis showed readily observable TOM+ human cells in the bone marrow (Fig. 4c), whilst flow cytometry analysis revealed the presence of erythroid, myeloid and B lymphoid cells in the bone marrow, as well as splenic B and T cells and, in some animals, developing thymic T cells (Fig. 4d–g, Extended Data Figs. 5b and 6a,b and Supplementary Table 13). Immature CD3− thymocytes passed from the CD4−CD8− stage through an intermediate single-positive CD4+ stage to CD4+CD8+ double-positive thymocytes and CD3+ double-positive thymocytes gave rise to single-positive CD4+ and CD8+ T cells (Fig. 4g, Extended Data Fig. 6b and Supplementary Table 13). Erythroid cells in the bone marrow stained for cell-surface GYPA and CD43 and predominantly expressed low levels of the TOM or BFP reporter genes (Fig. 4d and Extended Data Fig. 5b). This was consistent with prior observations that maturing erythroid cells preferentially transcribed globin genes and reduced expression from the GAPDH locus28,29. Another defining characteristic of the MLE animals was the presence of a bone marrow CD45+CD34+CD38lo/− HSC-like population (Fig. 4d, Extended Data Figs. 5b and 6a and Supplementary Table 13).
Modulating VEGF signaling enhances MLE in recipients from multiple independent iPS cell lines
The lack of efficient generation of iHSCs from the PB1.1 BFP cell line in cohort 3 mice led us to consider modifications to the protocol growth factor composition that might improve the robustness of hematopoietic differentiation. Evidence from the human embryo13 suggests that HSCs arise from an arterially patterned hemogenic endothelium. VEGF acts in a dose-dependent manner to drive endothelium generation and arterialization in differentiating PS cells30,31,32,33. However, recently published work using a murine embryonic stem cell differentiation system showed that VEGF suppressed hematopoietic progenitor development from endothelium by blocking the upregulation of Runx1 expression21, a critical marker of hemogenic endothelium and regulator of HSC development in the mammalian embryo13,34. To explore these opposing effects, we trialed a range of VEGF concentrations from days 3 to 7, during endothelial generation, followed by continuing or removing VEGF to determine which best enhanced the endothelial-to-hematopoietic transition. We demonstrated a VEGF dose-dependent increase in CD34+CXCR4+ arterial endothelial cell generation, followed by a rapid loss of the arterial marker CXCR4 after the removal of VEGF on day 7 of differentiation (Extended Data Fig. 7). Gene expression analysis revealed that the combination of high VEGF from day 3 followed by its removal on day 7 of differentiation increased the expression of aortic endothelial genes (AGTR2, IL33 and EDN1), reduced ALDH1A2 and increased ALDH1A1, accelerated the endothelial-to-hematopoietic transition, evidenced by the reduction in CXCR4 and DLL4, and increased RUNX1 and HLF expression (Extended Data Fig. 8). We previously identified many of these genes as being more lowly expressed in iPS cell-derived cells compared to the human embryo13 in an endothelial-to-hematopoietic transition scorecard (Extended Data Fig. 4f).
We incorporated these modifications into the next evolution of the differentiation protocol (denoted protocol 3; Extended Data Fig. 1a) and explored their functional consequences in further transplantation experiments. In mice transplanted with RM TOM cells (cohort 4), we observed improved engraftment compared to the earlier experiments (cohorts 1 and 2), recording 30/62 (48.4%) mice with MLE, with 61/62 recipients analyzed >16 weeks after transplantation (Fig. 5a and Supplementary Tables 2, 3 and 14). Similar engraftment results were seen in three additional human iPS cell lines, including the PB1.1 BFP line that transplanted less efficiently in experiments (cohort 3) using the previous differentiation protocol (Extended Data Fig. 1a). MLE was observed in 11/23 (47.8%) of PB1.1 BFP (cohort 5), 4/15 (26.7%) of PB5.1 (cohort 6) and 3/8 (37.5%) of PB10.5 (cohort 7) mice, analyzed >16 weeks after transplantation in 41/46 cases (Fig. 5b–d and Supplementary Tables 2, 3 and 15–17). These results indicated that protocol 3 generated more robustly engrafting cells and was applicable to a broader range of iPS cell lines. We have not yet performed limit dilution transplantation experiments using this protocol but analysis of the overall engraftment results given above suggests that the frequency of MLE cells was 1 in 3.0 × 106 for the RM TOM, 1 in 3.1 × 106 for the PB1.1 BFP, 1 in 6.2 × 106 for the PB5.1 and 1 in 4.3 × 106 for the PB10.5 lines35. We found some variability in outcomes between experiments, with estimated engraftment frequencies as high as 1 in 1.3 × 106 for RM TOM experiment E427 in which 7/9 recipients displayed MLE (Supplementary Tables 14–17). In summary, these results still show variation in engraftment between different lines and between experiments. We believe that this variability may be lessened with further protocol optimization.
a–d, Engraftment of BM and SPL in transplant recipients of RM TOM (a), PB1.1 BFP (b), PB5.1 (c) and PB10.5 (d) cells showing the phenotype of engrafting cells and the level of engraftment. Error bars, mean ± s.e.m. e–h, Tissue distribution of engrafting cells in MLE recipients of RM TOM (e), PB1.1 BFP (f), PB5.1 (g) and PB10.5 (h) cells in BM, SPL, THY and PB at 12 and 16 weeks. Error bars, mean ± s.e.m. i, Flow cytometry analysis of BM in engrafted mice for each cell line showing GYPA+ erythroid lineage and CD45+ lymphoid and myeloid cells. j, BM, SPL and THY or mediastinal lymph node (LN) tissue of RM TOM-engrafted mouse m574, showing GYPA+ erythroid, CD45+CD19+ B cell, CD45+CD3+ T cell, CD45+CD33+/CD13+ myeloid and CD45+CD34+CD38lo/- stem cell populations in the BM, CD45+sIgM+ B cells and CD45+CD3+ T cells in the SPL and THY or mediastinal LN tissue containing CD45+CD3+CD4+ and CD45+CD3+CD8+ T cells and a population of CD45+CD19+ B cells.
We analyzed the contribution and lineage distribution of human cells in the bone marrow, spleen, thymus and peripheral blood of the 48 MLE animals receiving cells differentiated under protocol 3 (Fig. 5e–h and Extended Data Fig. 9). In most cases, human cells were present in the peripheral blood at 12 weeks after transplantation (38/46 mice analyzed) (Fig. 5e–h) and the evaluation of paired samples at 16 weeks revealed an increase in human cells in 28/35 mice analyzed across the four cell lines (Extended Data Fig. 9a). There was an evident sex bias in bone marrow engraftment, most prominent in the RM TOM line recipients (Extended Data Fig. 9b), with significantly higher levels of human cells in female than male recipients, consistent with the published literature36. Over all experiments (cohorts 1–7), MLE was seen in 19.6% of recipients transplanted with CD34+ suspension blood cells, 23.8% of those receiving CD34-enriched cells from the EBs and 24.5% of mice that received both suspension blood cells and CD34-enriched cells from the EBs. These proportions were not statistically different and demonstrated that stem cells were present in CD34+ cells from both sources (Supplementary Table 18).
Flow cytometry analysis revealed the presence of bone marrow erythroid, myeloid, B and T lymphoid cells and CD45+CD34+CD38lo/− HSC-like cells, splenic B and T cells and thymic T cells, similar to MLE animals in cohorts 1–3 (Fig. 5i,j, Extended Data Fig. 9c–e and Supplementary Table 19). A comparison of mice engrafted with RM TOM cells under the different protocols revealed higher percentages of human cells in the bone marrow, spleen and peripheral blood in recipients of protocol 3 differentiated cells, with a persistent bias toward higher engraftment in female mice (Supplementary Fig. 4a,b). The proportions of erythroid, myeloid, B and stem cells were similar in male and female recipients but the proportion of T cells in the bone marrow and spleen were greater in engrafted female mice (Supplementary Fig. 4c,d).
Where T cell development was observed in the spleen and bone marrow, we rarely observed a macroscopically identifiable bilobed thymus but small amounts of putative lymphoid tissue were frequently present in the mediastinum. This tissue usually contained single-positive CD4+ and CD8+ cells, together with few double-positive thymic cells and often a population of CD19+ B cells (Fig. 5j, Extended Data Fig. 9e and Supplementary Table 19). The low percentage of CD4+CD8+ cells was particularly marked in the more highly engrafted cohort 4 female (1.8% ± 0.7% CD4+CD8+ cells) compared to male (23.7% ± 6.9% CD4+CD8+ cells) mice receiving cells differentiated using protocol 3 and contrasted with the high proportion of CD4+CD8+ cells seen in female (52.7% ± 12.1%) and male (75.7% ± 4.1%) mice receiving protocol 1 and 2 differentiated cells (Supplementary Fig. 4c,d and Supplementary Table 20). We speculate that this result might reflect an inability to sustain thymic tissue in aging immune-deficient mice, with also likely sampling of mediastinal lymph nodes to account for the presence of B cells (Fig. 5j and Supplementary Tables 19 and 20). These observations may have been more prominent in cohort 4 recipients because the degree of T cell engraftment, evident by T cell contribution to the human cells in the bone marrow and spleen, was greater in recipients of protocol 3 differentiated cells (compare cohort 4 recipients in Fig. 6a to cohort 1–3 recipients in Extended Data Fig. 10a). A similar dimorphic pattern of T cell engraftment was observed in immune-deficient mouse recipients of CB CD34+ cells, in which a major CD4+CD8+ thymic population was seen in 10/19 mice with T cell engraftment, while low CD4+CD8+ cell numbers were seen in the remainder37.
a, Top: bar graphs showing the level of human engraftment in the BM of MLE mice receiving the indicated cell lines (individual recipients identified on x axis). Bottom: stacked column graphs showing the lineage distribution of human cells in the BM of iHSC-engrafted recipients. UN, unclassified cells include myeloid, dendritic and natural killer cells not detected by the antibodies used (Supplementary Table 19). b–h, Characteristics of engrafted CB cells. b, Scatter plot correlating calculated dose of injected CD34+ CB cells with phenotype and level of human engraftment in the BM. Each circle represents one animal, color-coded to represent M, ML, ME and MLE patterns of engraftment. Error bars, mean ± s.e.m. A total of 39 animals were transplanted. c, Flow cytometry plot showing GYPA+ erythroid cells and CD45+ lymphoid and myeloid cells. d, Tissue distribution of engrafting cells in MLE recipients of CB cells in BM, SPL, THY and PB at and 16 weeks. e, Analysis of paired samples of PB showing increased levels of human cells in 6/8 recipients between 12 and 16 weeks. f,g, Lineage distribution in the BM (f) and SPL (g) in CB recipients. h, Top, bar graphs showing the level of human engraftment in BM of MLE mice receiving CB cells (individual recipients identified on x axis). Bottom, stacked column graphs showing the lineage distribution of human cells in the BM of CB-engrafted recipients. UN, unclassified cells include myeloid, dendritic and natural killer cells not detected by the antibodies used (Supplementary Table 21).
Heterogeneity of lineage composition in MLE recipients of iHSCs
Despite robust overall transplantation of human cells, lineage contributions varied among different MLE mice and recipients of cells differentiated from independent iPS cell lines (Extended Data Figs. 6 and 9 and Supplementary Tables 13 and 19). The bone marrow of most mice receiving RM TOM cells in cohorts 1–3 was dominated by B cells (Extended Data Fig. 10a). Some mice showed predominantly erythroid engraftment and the cohort 4 mice receiving cells differentiated under protocol 3 also frequently displayed T cell engraftment (Fig. 6a). Recipients of PB1.1 BFP differentiated cells (cohort 5) displayed dominant erythroid engraftment, whilst the smaller number of recipients of PB5.1 (cohort 6) and PB10.5 (cohort 7) lines showed more balanced engraftment patterns (Fig. 6a, Extended Data Fig. 9 and Supplementary Table 19). We observed that engraftment was maintained in the bone marrow and spleen in animals evaluated for >16 weeks, consistent with stable engraftment by long-term repopulating cells (Supplementary Fig. 5).
Umbilical CB mononuclear cells display dose-dependent engraftment
We wished to provide a relevant context for our experiments by comparing the engraftment phenotypes of iPS cell-derived iHSCs to those of CB cells, a clinically validated source of repopulating HSCs. A total of 39 mice were transplanted with 5 × 104–2.5 × 106 CB mononuclear cells isolated from four separate cords that comprised 0.7–2.7% CD34+ cells, resulting in the transplantation of 3.5 × 102–2.7 × 104 CD34+ cells. MLE was observed in most recipients (14/15) of mononuclear cells calculated to contain > 6.0 × 103 CD34+ cells (Fig. 6b and Supplementary Table 21) and the estimated frequency of repopulating CB stem cells was 1 in 6.3 × 103 CD34+ cells according to a limit dilution assay35 (Extended Data Fig. 10b), consistent with reports in the literature37. Similar to our findings with iPS cell-derived iHSC transplants, CB cells showed higher levels of engraftment in female mice (Extended Data Fig. 10b,c). Mice receiving fewer than 6.0 × 103 CB CD34+ cells showed lower total proportions of human cells in the bone marrow. Moreover, they frequently displayed restricted lineage engraftment with myeloid or myeloid and lymphoid lineages (Fig. 6b). This positive correlation between engraftment level and MLE in recipients of CB stem cells mirrored the similar correlation observed in the iPS cell-derived blood cell transplants (Supplementary Results 2). This observation also aligned with reported dose-dependent hematopoietic chimerism in immune-deficient mice receiving purified CB stem cells, where engraftment with low stem cell numbers similarly led to low levels of myeloid or myeloid-restricted and B cell-restricted engraftment that persisted for 19–21 weeks37. Taken together, this indicated that the transplantation assay reads out a hierarchy of stem cells for both CB-derived and iPS cell-derived cells. MLE cells with high proliferative capacity are less abundant than myeloid or myeloid-restricted and lymphoid-restricted stem cells with low proliferative capacity.
The profile of engrafted lineages was similar between CB and iHSC recipients, although T cell engraftment was greater in the RM TOM mice (compare Fig. 6c–g to Extended Data Fig. 9a–d). PB1.1 BFP recipients displayed prominent erythroid engraftment in the bone marrow and spleen, with commensurately lower levels of lymphoid and myeloid engraftment (Extended Data Fig. 9c–d). In CB recipients, the most abundant lineages were B and myeloid cells, with few mice displaying large erythroid populations and few cases of T cell engraftment. Heterogeneity in the distribution of bone marrow lineages in individual MLE CB mice can be appreciated in the bar graphs in Fig. 6h and compared to similar data for iHSCs shown in Fig. 6a and Extended Data Fig. 10a.
iHSCs show comparable secondary engraftment to CB HSCs
We investigated whether bone marrow cells from primary recipients engrafted with either CB-derived or iPS cell-derived HSCs could engraft secondary recipients. We observed secondary engraftment from 6/12 primary mice engrafted with iHSCs and from 2/5 primary mice engrafted with CB cells, with similar outcomes observed from primary recipients engrafted with cells generated by different protocols (Discussion, Supplementary Discussion and Supplementary Table 22). In most cases, engraftment was at a low level and restricted to myeloid lineages, although one iHSC secondary transplant recipient displayed B, T and myeloid lineages in the bone marrow, spleen and thymus.