Cell lines
Healthy donor, FANCA–/– (FA55) and FANCD2–/– (FA-75) LCLs were generously gifted by Paula Rio (CIEMAT). K562, Jurkat, U2OS, MD-MBA-431, RPE-1 hTERT and HeLa cells were obtained from the American Type Culture Collection or the Berkeley Cell Culture facility. The RPE-1 hTERT TREX1−/− cell line was used in this study32. LCLs were cultured in RPMI 1640 medium (GlutaMAX, from Thermo Fisher Scientific, 61870010) supplemented with 20% Gibco FBS (Thermo Fisher Scientific, 10270106), 1% Gibco penicillin–streptomycin (P/S) solution (Thermo Fisher Scientific, 15140122), 0.005 mM Gibco β-mercaptoethanol (Thermo Fisher Scientific, 31350010) and 1% Gibco MEM non-essential amino acids (Thermo Fisher Scientific, 11140050). K562 and Jurkat cells were cultured in RPMI 1640 GlutaMAX media supplemented with 10% FBS and 1% P/S solution. U2OS, MD-MBA-421, RPE-1 hTERT and HeLa cells were cultured in DMEM, high glucose, GlutaMAX pyruvate medium (Thermo Fisher Scientific, 10569010) supplemented with 10% FBS and 1% P/S. All cells were cultured at 37 °C with 5% CO2 in a humidified incubator. Cell lines were regularly tested for mycoplasma with a MycoAlert Mycoplasma Detection Kit (Lonza, LT07-318).
To generate CRISPRi cell lines, we packaged a CRISPRi (pHR-EF1a–dCas9–HA–mCherry–KRAB–NLS) construct into lentiviruses in HEK293T cells. The lentiviral supernatant was filtered and used to transduce LCLs and HeLa cells. After transduction, mCherry+ cells were sorted using an SH800 Cell sorter (Sony). Because LCLs failed to survive as single cells in 96-well plates, we first seeded approximately 500 mCherry− LCLs per well in 96-well plates and then sorted single mCherry+ cells into 500 mCherry− cells to overcome the viability problem. mCherry+ LCLs were further enriched with consecutive rounds of sorting until reaching mCherry purity higher than 90%.
For retroviral transduction of 3×flag-TREX1-wt in RPE-1 hTERT TREX1−/− cells, open reading frames were cloned into pQCXIZ, which confers resistance to zeocin. Constructs were transfected into Phoenix amphotropic packaging cells using calcium phosphate precipitation. Cell supernatants containing retrovirus were filtered, mixed 1:1 with target cell media and supplemented with 4 μg ml−1 polybrene. Successfully transduced cells were selected using zeocin (Life Technologies).
Primary cell culture
CD34+ HSPCs were cultured in SC media (SFEMII and CC110 (STEMCELL Techonologies)). CD4+ T cells were purified from frozen human peripheral blood Leukopak (STEMCELL Techonologies) by negative selection using an EasySep Human T Cell Enrichment Kit (STEMCELL Technologies) according to the manufacturer’s instructions and cryopreserved in CryoStor CS5 (STEMCELL Technologies). Purified T cells were cultured in X-VIVO 15 Media (Lonza) supplemented with 5% human AB serum (GeminiBio) and 100 IU ml−1 human IL-2 (Miltenyi Biotec). For gene editing experiments, T cells were activated using TransAct (Miltenyi Biotec) according to the manufacturer’s instructions. For Fig. 4e,f, T cells were activated 1 d after thaw using CD3/CD28 Dynabeads (Thermo Fisher Scientific) according to the manufacturer’s instructions. The beads were removed after 2 d. The T cells were used in gene editing experiments on either day 3 or day 4. iPSCs were cultured in complete StemFlex media (Gibco Life Technologies) and seeded in the coated plates with Synthemax (Corning).
Mouse cell lines
4T1 and CT26 cell lines were cultured in RPMI media (supplemented with 10% FBS and 1% P/S solution). E0771.LMB cells were cultured in DME HG media (supplemented with 10% FBS, 5% HEPES and 1% P/S solution). All cells were cultured at 37 °C with 5% CO2 in a humidified incubator.
In vitro transcription of gRNAs
gRNAs were in vitro transcribed as described (https://doi.org/10.17504/protocols.io.dwr7d5)57. In brief, overlapping oligomers, indicated in Supplementary Table 2, containing a T7 promoter, protospacer and gRNA scaffold, were amplified by Q5 High-Fidelity DNA Polymerase (New England Biolabs, M0491L) for 15 cycles. Then, 1 µM T7FwdLong and 1 µM T7RevLong were used as a template and amplified by T7FwdAmp and T7RevAmp in 50-µl reaction volume. Next, 8 µl of PCR-amplified product was used for the in vitro transcription using an NEB HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs, E2040S), incubating at 37 °C for 18 h in the thermocycler. Then, the reaction was supplemented with DNase I (Qiagen, 79256) for 30 min at 37 °C, followed by Quick CIP (New England Biolabs, M0525S) treatment for 1 h at 37 °C. The gRNAs were later purified with an miRNeasy kit (Qiagen, 217604), and concentration was measured by Qubit RNA Broad Range assay (Thermo Fisher Scientific, Q10211) and stored at −80 °C.
Genome-wide library construction
To shuttle genome-wide sgRNAs, we first amplified the cassettes including sgRNA using the primer set priEK-35 and priEK-37 from the CRISPRi-V2 library (Addgene, 1000000093) using Phusion polymerase (New England Biolabs, M0530L) under the following condition: 30 s at 98 °C, then 15 cycles of 15 s at 98 °C, 15 s at 53 °C, 15 s at 72 °C and then a final extension for 10 min at 72 °C. Amplified PCR fragments were digested overnight with Bpu1102I (BlpI) (Thermo Fisher Scientific, ER0091) and BstXI (Thermo Fisher Scientific, ER1021) at 37 °C. The digested DNA fragments were separated in a 10% TBE gel to cut the DNA band corresponding (~33 base pairs (bp)). The gel pieces were crushed by spinning for 3 min at 20,000g and then eluted in water at 37 °C overnight. DNA later was precipitated with the NaOAc/EtOH method. In addition, we linearized the vector carrying mutated GFP sequence with the same restriction enzymes, Bpu1102I (BlpI) and BstXI, for 4 h at 37 °C. The linearized DNA product was separated by 0.8% agarose gel electrophoresis and excised from the gel. DNA was cleaned using a QIAquick Gel Extraction Kit (Qiagen). The DNA further was cleaned with the NaOAc/EtOH method. For the ligation reaction, 500 ng of linearized vector and 1.9 ng of insert were incubated with T4 DNA Ligase (New England Biolabs, M0202L) for 16 h at 16 °C. The ligated plasmids were purified by isopropanol/5 M NaCl precipitation and resuspend in 13 µl of elution buffer. Then, 1 µl or 2 µl of the purified ligation reaction was mixed with 25 µl of MegaX DH10B T1R electrocompetent cells (Thermo Fisher Scientific, C640003) and recovered in S.O.C medium for 1.5 h. The bacteria were plated in 24.5 × 24.5-cm LB agar plates containing ampicillin resistance. While plating the bacteria in 24.5 × 24.5 cm, the dilutions of bacteria were performed as well to detect approximate coverage of the sgRNA library. Grown colonies were collected by scraping from LB agar plates. Plasmids were recovered by several midi-preps (Qiagen Plasmid Plus Midi Kit).
Quality of the library was determined by NGS. Sequencing libraries were prepared by amplifying sgRNA cassettes with the primers priEK_i5-1 and priEK_i7-1 and secondary PCR to put the sequencing adapters priEK_501 and priEK_701 (indicated in Supplementary Table 2). The reaction was sequenced by MiSeq, and sgRNA distribution of the cloned library was analyzed using custom scripts.
Production of sgRNA library lentiviruses
To produce lentivirus from the sgRNA library, approximately 7 million HEK293T cells were seeded in a 15-cm plate in 20 ml of DMEM medium with 10% FBS and 1% P/S. The next day, HEK293T cells were transfected with the library. Per plate, in a 5-ml tube, 15 µg of sgRNA library, 12 µg of delta VPR and 3 µg of VSVG were resuspended with 1.3 ml of Opti-MEM and mixed with 270 µl of polycation polyethylenimine (PEI) 1 mg ml−1 (1 µl to 3 µg of DNA). The mix was incubated at room temperature for 20 min and then added on top of HEK293T cells in a drop-wise manner. The media were changed on the following day. The virus-containing cell culture media were collected 48 h and 72 h after the transfection. The viral media were combined and filtered using a 0.45-µm PES membrane (Thermo Fisher Scientific, 295-3345), aliquoted into 15-ml Eppendorf tubes, snap frozen and stored at −80 °C.
CRISPR screen
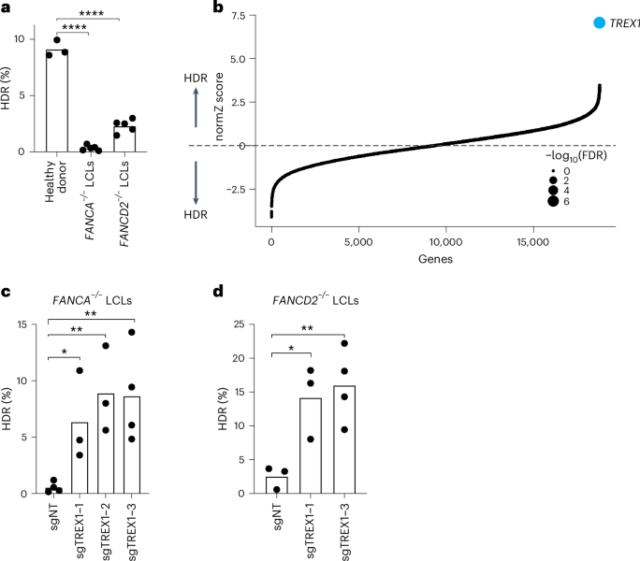
FANCA–/– CRISPRi cells were grown to 150 million cells before transduction with the genome-wide CRISPRi library. Because LCLs were extremely difficult to transduce with lentiviruses, FANCA–/– CRISPRi cells were directly resuspended in virus-containing media and seeded in six-well plates in the presence of 8 µg ml−1 polybrene. The coverage determined by BFP+ cells was around 300× per sgRNA. Twenty-four hours after transduction, cells were collected and transferred to T75 flasks. A day later, gRNA-containing cells were selected with puromycin treatment (0.5 µg ml−1) for 96 h. At this moment, BFP+ cells were over 90%. Upon puromycin selection achieved, cells were split into two replicates and were maintained for 250× coverage throughout the screen. Twenty days after transduction, cells were subjected to Ficoll gradient (Ficoll Paque Plus, Millipore Sigma, GE17-1440-02). In total, 1 × 106 cells were electroporated with 400 pmol SpCas9 nuclear localization sequence (NLS), 480 pmol L2 gRNA targeting BFP and 500 pmol BFP-to-GFP ssODN template using CM-189 and SF solution (Lonza, 4D electroporator) per replicate. Cells were further expanded in culture before sorting. Before the sort, a background population was collected for downstream NGS analysis. The sort was performed in two steps: first, thresholding was set to enrich the GFP+ population from approximately 0.5% to 70%, and then a stringent sort was performed to achieve approximately 99% pure GFP+ cells. Cells were pelleted and stored at −80 °C until genomic DNA (gDNA) extraction.
NGS sample preparation and screen analysis
gDNA was extracted using a Gentra Puregene Cell Kit (Qiagen, 158912) gDNA extraction protocol. In brief, for the background samples (from a total of 25 million cells), the cell pellet was resuspended in 3 ml of cell lysis solution and then mixed with 15 µl of RNase A solution at 37 °C for 20 min and cooled down for 10 min on ice, and then 1 ml of protein precipitation buffer was added. The mix was vortexed thoroughly and spun down for 10 min at 2,000g. The gDNA-containing supernatant was mixed with 100% isopropanol (3 ml) by inverting the 15-ml tube for 50 times. The gDNA was pelleted by centrifugation at 2,000g for 5 min and then washed with 70% EtOH. After removing 70% EtOH, gDNA was resuspended in 200 µl of hybridization buffer and incubated at 50 °C for 1 h. DNA amount was measured by NanoDrop. For the GFP+ cells, the protocol was adjusted for the low cell number. In the gDNA precipitation step, glycogen was added to facilitate the gDNA precipitation.
Purified gDNA was used for the further PCR amplification of sgRNA cassettes following the protocol (https://weissman.wi.mit.edu/resources/IlluminaSequencingSamplePrep.pdf). For the background samples, 5 µg of gDNA per reaction was used. For the sort background samples, 30 PCR reactions were performed and combined later. Because we had very limited DNA from GFP+ cells, we used 1-µg amount of DNA per reaction and performed two reactions. The PCR products were purified by two rounds of Sera-Mag magnetic beads (Cytiva, 29343957). The concentrations were measured by a Qubit 1× dsDNA High Sensitivity Assay (Thermo Fisher Scientific, Q33232), and the samples were pooled according to their anticipated read counts. The samples were sequenced on a NextSeq 2000.
Screening data were analyzed using the standard protocols in MaGECK (version 0.5.9.5) and drugZ. MaGECK was used to get the gRNA counts per each sgRNA in the population by using the following command:
mageck count -l CRISPRi_v2_humantop5.library.txt–fastq 20210427A-EK1_R1.fastq.gz 20210427A-EK2_R1.fastq.gz 20210427A-EK3_R1.fastq.gz 20210427A-EK4_R1.fastq.gz 20210427A-EK5_R1.fastq.gz 20210427A-EK6_R1.fastq.gz–sample-label bgsort_rep1,bgsort_rep2,bg_rep1,bg_rep2,gfp_rep1,gfp_rep2 -n count_mageck.txt
drugZ was used to integrate multiple guides into gene-level phenotypes relative to the background unsorted population (normZ score and FDR values)23,24 by using the command:
python drugz.py -i count_mageck.txt -o drugz-gfpposvsbgsort.txt -c bgsort_rep1,bgsort_rep2 -x gfp_rep1,gfp_rep2
(Supplementary Table 1).
ggplot in R was used to produce the graph in Fig. 1b by using the following code:
drugz_gfp_sort <- read_tsv(“~/Documents/FALCLScreen_results/drugZ_output/drugz-gfpposvsbgsort.txt”)
ggplot(drugz_gfp_sort, aes(y = normZ, x = rank_synth, size = -log10(fdr_supp)))+
geom_jitter(alpha = 0.4)+
theme_classic()
ggsave(“drugZ_v2.pdf”,width = 20, height = 10, units = c(“cm”))
Further modifications were made using Adobe Ilustrator.
RNP electroporation for BFP-to-GFP reporter assay and genomic loci targeting
RNP electroporation was performed as described (https://doi.org/10.17504/protocols.io.dm649d)9. In brief, 36 pmol sgRNA and 30 pmol SpCas9-NLS were mixed in Cas9 buffer (20 mM HEPES at pH 7.5, 150 mM KCl, 1 mM MgCl2, 10% glycerol and 1 mM tris (2-carboxyethyl) phosphine (TCEP) reducing agent). The mixture was incubated at room temperature for 20 min. Then, 1 × 105 to 2 × 105 cells were collected and spun down at 300g for 5 min. The cell pellets were resuspended in 15 µl of nucleofection buffer (Lonza). Then, 5 µl of RNP mixture was added to the cell suspension with 0.3 µl of 100 µM (30 pmol) ssODN (BFP-to-GFP template) template. Five days after electroporation, cells were collected and subjected to flow cytometry with an Attune Flow Cytometer (Thermo Fisher Scientific). Downstream analysis was performed using FlowJo version 10.8.2 software.
Mouse cells were electroporated following the same protocol as other cells (see above) using the supplemented SE buffer provided by Lonza and using the CM-150 program.
For endogenous locus targeting, 100 pmol SpCas9-NLS was mixed with 120 pmol gRNA in Cas9 buffer, and the mixture was incubated for 20–30 min at room temperature or 37 °C. In total, 1 × 105 to 2 × 105 cells were collected and resuspended in 15 µl of nucleofection buffer (Lonza). For each reaction, 100 pmol ssODN was then added before nucleofections. Electroporations were performed in the strip format, with 20-µl volume of cells and RNP mix. The following kit and program for each cell type was selected: K-562 (SF kit/FF-120), RPE-1 (P3 kit/EA-104), U2OS (SE kit/CM-130), MDA-MB-231 (SE kit/CM-130), HeLa (SE kit/CM-130) and Jurkat (SE kit/CL-120). After electroporation, pre-warmed 80 µl of DMEM or RPMI medium was added into strips. Cells were incubated in the hood for 10 min and then transferred to the plates and returned to 37 °C.
ssODNs were purchased from Integrated DNA Technologies as Ultramer DNA oligos. To protect ssODNs, they were ordered with four phosphorothioate modifications at the 5′ and/or 3′ ends. To amplify PCR templates, we performed with primers for the corresponding genes following the PCR protocol: 30 s at 98 °C, then 15 cycles of 15 s at 98 °C, 15 s at 60 °C, 15 s at 72 °C and then a final extension for 10 min at 72 °C. We later used Sera-Mag beads to clean and concentrate the PCR products around 700–1,000 ng µl−1 and used 1,000 ng of template per nucleofection. Oligo sequence information can be found in Supplementary Table 2.
For the primary cell nucleofection, the protocol described above was used with the following changes. For HSPCs, cells were electroporated with P3 kit, ER-100 or DS-130 program. Activated T cells were electroporated with P3 kit and CN-114 program, and iPSCs were electroporated with P3 kit and CB-150 program. After electroporation, cells were returned to their corresponding media, as indicated above.
For rAAV donors, custom-made ssAAV vectors with serotype 6 were prepared and sent to the viral vector facility (VVF) of the Neuroscience Center of Zurich (ZNZ) to generate viral particles. AAVs were ultracentrifuged (OptiPrep) and dial-filtered in 1× PBS pH 7.4, 1 mM MgCl2 and 2.5 mM KCl. The titers of the donors were determined as 7.9 × 1012 vector genomes per milliliter (vg/ml) and 8.1 × 1012 vg/ml, respectively. Approximately 1010 viral particles for 1 × 105 cells were added 5 min after nucleofection. The media were changed either after overnight or 2 d after transduction.
gDNA extraction
Cell pellets were collected 72–96 h after electroporation and resuspended in QuickExtract solution (Lucigen, QE09050) and subjected to gDNA extraction while incubating for 10 min at 65 °C, 5 min at 98 °C and then holding at 4 °C. After the incubation, 1 µl of gDNA was taken for further PCR reactions for NGS.
NGS
Primers containing adaptor binding sites (indicated in Supplementary Table 2) were designed to amplify 150–200 bp around the cut sites. First, gDNA was amplified using NEBNext Ultra II Q5 Master Mix for 30 cycles and then cleaned with SPRI beads (SeraMag Select (Cytiva, 29343052) or in house). From the purified reactions, around 10–20 ng of DNA was used as input for the second PCR reaction to add i7/i5 indexes for the samples in nine reaction cycles. Amplicons were then purified again with 0.8× SeraMag beads, and samples for the same genomic loci were combined. The amplicon length and purity were analyzed by a TapeStation with D1000 DNA flow cells (Agilent). Pools were combined based on their amount and desired read number (50,000–100,000 reads per sample). The combined samples were sequenced in Illumina sequencers (MiSeq or NextSeq 2000) in the Genome Engineering and Measurement laboratory at ETH Zurich.
NGS analysis
The sequencing reads were demultiplexed and analyzed with CRISPresso2 (version 2.0.20b) in batch mode58 with default parameters other than minimum average read quality (- q) of 30 and minimum single bp quality of (- s) 10 and the quantification window (- w) 20. Reads with a frequency lower than 0.5% were disregarded before further analysis. Results were then normalized to sum up to 100%.
Immunoprecipitation–western blot
Cells indicated in Figs. 2, 4 and 5 were harvested by trypsinization and lysed in RIPA buffer (5 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS), supplemented with phosphatase inhibitors (10 mM NaF, 20 mM β-glycerophosphate) and protease inhibitor (Thermo Fisher Scientific) at approximately 106 cells per milliliter and incubated on ice for 20 min. Lysates were sonicated with a Bioruptor Plus sonication device (Diagenode) for 15 cycles ON/OFF (high, 4 °C). Sonicated lysates were incubated on ice for 20 min and centrifuged at 16,000g and 4 °C for 20 min, and supernatants were transferred into French tubes before protein quantification using the Pierce BCA protein assay (Thermo Fisher Scientific). Lysate equivalent to 10–50 µg of proteins was mixed to 1× Laemmli buffer (50 mM Tris, 10% glycerol, 2% SDS, 0.01% bromophenol blue, 2.5% β-mercaptoethanol), resolved by SDS-PAGE (Life Technologies) and transferred to nitrocellulose membranes (Amersham). Membranes were blocked in 5% milk in TBS with 0.1% Tween 20 (TBS-T) and incubated with primary antibody overnight at 4 °C, washed three times in TBS-T and incubated for 1 h at room temperature with HRP-conjugated secondary antibody. After three washes in TBS-T, imaging was performed using enhanced chemiluminescence (Thermo Fisher Scientific).
Antibodies
Primary antibodies included: anti-TREX1 (1:1,000 dilution) (Abcam, ab185228), anti-β-actin (1:2,000 dilution) (Abcam, ab8224), anti-RPA32 (1:1,000 dilution) (Abcam, ab2175), anti-flag (1:2,000 dilution) (Abcam, f1804) and anti-HSP60 (1:1,000 dilution) (Santa Cruz Biotechnology, sc-1052).
Secondary antibodies (1:10,000–15,000 dilution) included: goat anti-mouse IgG HRP (Thermo Fisher Scientific, 31432), donkey anti-rabbit IgG HRP (SouthernBiotech, 6441-05), anti-rabbit secondary antibody IRDye 800 CW (LI-COR Biosciences, 926-32213) and anti-goat IRDye 800 CW (LI-COR Biosciences, 926-32214).
Biotin-ssODN immunoprecipitation
In total, 5–6 × 106 RPE-1 hTERT parental or TREX1−/− cells expressing were electroporated with 5 nmol biotinylated or unprotected ssODN (Integrated DNA Technologies) in 100 µl of final volume using a Lonza 4D-Nucleofector (Methods). Two hours after electroporation, cells were harvested by trypsinization, washed with PBS and resuspended in lysis buffer (50 mM Tris pH 7.5, 200 mM NaCl, 0.075% NP-40, protease inhibitors) at 107 cells per milliliter. Cells were then dounce homogenized by 10 strokes with a tight-fitting pestle. Lysates were incubated on ice for 20 min and centrifuged at 16,000g and 4 °C for 20 min, and then input samples were taken for immunoblotting. To reduce non-specific binding of proteins to the beads, lysates were precleared by incubation with Protein G Dynabeads (Invitrogen) for 30 min at room temperature. To pull down the biotinylated ssODN, the cleared lysate was transferred onto streptavidin Dynabeads (10 μl per sample; Invitrogen) and again incubated for 30 min at room temperature. The beads were washed eight times with lysis buffer and then eluted with 2× Laemmli buffer (100 mM Tris, 20% glycerol, 4% SDS, 0.02% bromophenol blue, 5% β-mercaptoethanol). Immunoblotting of input and eluted samples (diluted 1:2) was performed as described in the Methods.
qRT–PCR
RNA extraction was performed using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Then, 1 μg of RNA per sample was used for reverse transcription using iScript Reverse Transcription Supermix for qRT–PCR (Bio-Rad, 1708841) according to the manufacturer’s instructions. qRT–PCR reactions were set up using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, 1725271) and run in triplicates using a QuantStudio 6 system (Thermo Fisher Scientific). A complete list of primers used in qRT–PCR can be found in Supplementary Table 2.
Imaging of TREX1-GFP and H2B-iRFP during gene editing
In total, 700,000 RPE hTERT GFP-TREX1 H2B-iRFP cells were nucleofected with Cas9 RNP (final concentrations: sgNT (L2 sgRNA) or CXCR sgRNA 0.72 μM, Cas9 enzyme 20 μM) with or without ssODN templates (BFP ssODN or CXCR4 template ssODN, final 5 μM). The nucleofected cells were plated on poly-lysine-coated chambered coverslips (ibidi, 80807) and imaged 24 h after nucleofection.
Live-cell imaging was performed at room temperature using a Nikon Eclipse Ti2-E equipped with a CSU-W1 SoRa spinning disk super-resolution confocal system, a Borealis microadapter, a Perfect Focus 4, a motorized turret and encoded stage, a five-line laser launch (405 (100 mW), 445 (45 mW), 488 (100 mW), 561 (80 mW), 640 (75 mW)), a Prime 95B monochrome digital camera and a CFI Apo TIRF ×60/1.49 NA objective lens. Images were acquired using NIS-Elements Advanced Research Software on a Dual Xeon Imaging workstation.
All images were processed by manually isolating the most in-focus single z-slice using ImageJ software. After separating channels using the ImageJ Macro ‘Batch Split Channels’ tool, images were processed by customized CellProfiler modules, where primary nuclear masks were generated, and the perinuclear ring was defined as the nuclear periphery to serve as a proxy for cytoplasm. The nuclear intensity of both GFP-TREX1 and H2B-iRFP channels were divided by perinuclear ring intensity and log10 transformed to provide log10(ratio of nuclear/cytoplasmic GFP-TREX1 or H2B-iRFP).
To access co-localization between GFP-TREX1 and H2B-iRFP channels, both channels of each image were subjected to the ImageJ plugin JACoP with manually adjusted threshold. Pearsonʼs coefficients and Mander’s coefficients were obtained, with M2 representing the fraction of GFP-TREX1 overlapping with H2B-iRFP.
Gene editing efficiencies at CXCR4 loci were analyzed as described above by NGS.
Statistical analysis
Each point represents an individual biological replicate, and bars represent the mean of the replicates. All P values were calculated using an unpaired t-test, *P < 0.05, **P < 0.01, ***P < 0.001, using GraphPad Prism version 9.4.1 software.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.