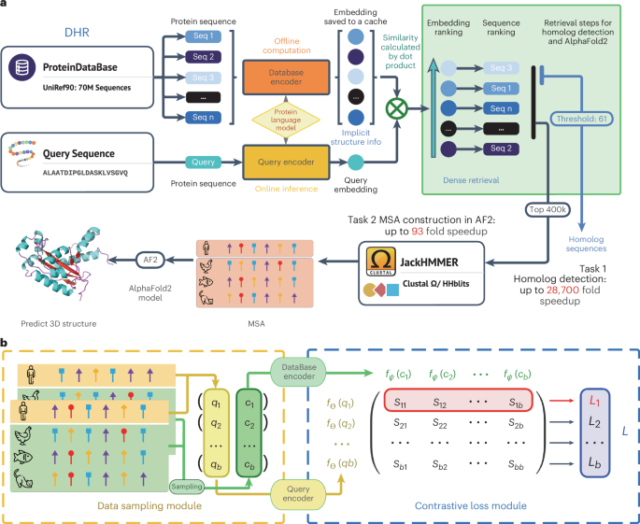
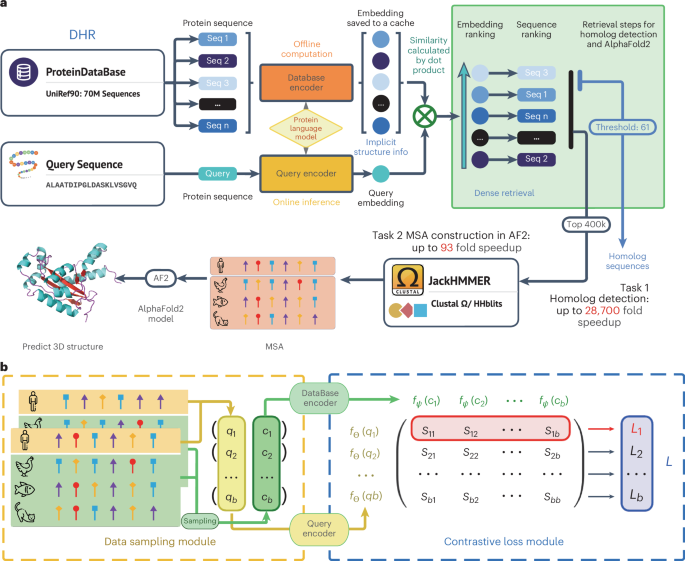
An ultrafast and sensitive protein homolog searching pipeline
The main idea underlying the proposed pipeline to detect homologs is to encode the protein sequences into dense embedding vectors and the similarity among sequences can be computed in a very effective manner. To be more specific, we empowered our method with a protein language model by initializing with ESM23,24 and integrating the contrastive learning technique as a way to effectively train our sequence encoder (database encoder and query encoder). This allows DHR to incorporate rich coevolutionary and structural information24,25,31,32 with great efficacy in retrieving homologs. Our dual-encoder architecture then offers greater flexibility in our model, allowing the same protein sequence to generate different embeddings depending on its role as a query or candidate sequence (Fig. 1a). Contrastive learning then seeks to learn encoder transformations that embed positive input pairs nearby while pushing negative pairs far apart (Fig. 1b). Following the completion of the training phase in our dual-encoder framework, we are able to generate offline protein sequence embeddings of high quality.
a, We leveraged a three-level hierarchical structure for our new framework—DHR. The main focus during training is to obtain the protein database encoder and query encoder. Through offline inference on the protein database and caching of embeddings thereafter, we are able to carry out vector-level similarity calculations on the basis of our set query representation, allowing us to generate an ordering for these sequences. For two consecutive downstream tasks, the inference pipeline uses the dot product to acquire the K most related sequences. Subsequently, JackHMMER is used on this small retrieved dataset to construct an MSA for further tasks such as 3D structure prediction or protein function forecasting. Before running retrieval, it is possible to encode the UniRef90 into vectors offline without compromising the speed of building MSA. b, Training pipeline for the proposed method. In the data sampling module, a batch of positive pairs, (q1, c1), …, (qb, cb), are sampled from MSAs. Then, a similarity matrix S is computed using the contrastive loss, in which each element \({s}_{{ij}}={f}_{\Theta }{\left({q}_{i}\right)}^{t}{f}_{\Psi }({c}_{j})\). Intuitively, the diagonal of the similarity matrix is trained to be larger than the corresponding off-diagonal elements in the same row because they represent all of the positive pairs.
We then leverage these embeddings and similarity search algorithms to retrieve homologs for each query protein (Fig. 1a). By designating similarity as the retrieval metric, we are able to find similar proteins in a much more accurate fashion than with traditional methods and use the similarity between two proteins for further analysis. Then, JackHMMER is incorporated to construct an MSA for our retrieved homologs. Our rapid and effective technique of discovering homologs, named DHR, harnesses dense retrieval to simplify searching within the protein sequence embeddings without spending excessive time on progressive alignment or dynamic programming that is used in conventional methods. Our alignment-free approach is faster compared to alignment-based methods by avoiding the need to compute pairwise alignment scores over large datasets, with higher accuracy in comparison (Fig. 1a). Moreover, with great efficiency compared to AF2 default MSA construction, DHR can produce diverse MSAs that contain more information when considering their Meff (Eq. (2)). Such information has been proven to be beneficial in protein 3D structure modeling: AF2 with DHR MSA inputs may achieve comparable results to AF2 with default MSA settings. These results suggest that this method can act as an effective and efficient replacement for existing MSA pipelines. Furthermore, diverse MSAs from DHR can complement existing pipelines. We developed a hybrid model, DHR-meta, combining DHR and AF2 default, which outperformed both individual pipelines on CASP13DM (domain sequence) and CASP14DM targets. Additionally, DHR can scale to search large databases such as BFD/MGnify, producing high-quality MSAs33,34,35,36. These MSAs enhance protein structure prediction, rivaling ColabFold’s performance in the CASP15 contest and the New-Protein dataset.
Highly sensitive homolog extraction with structure-aware information
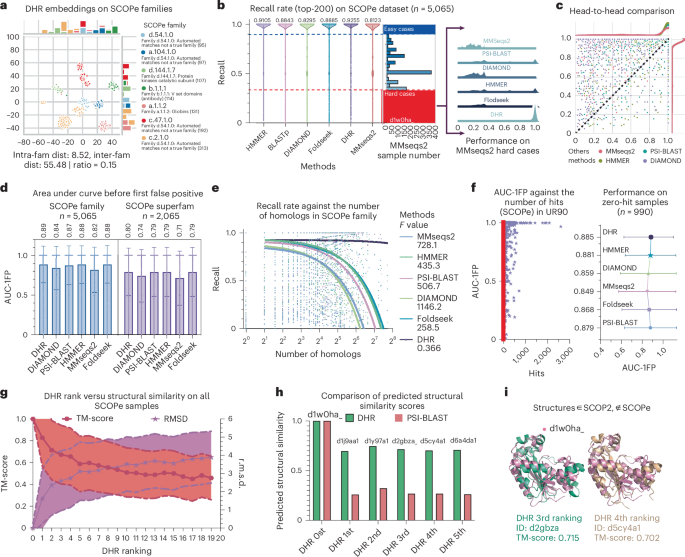
Having obtained the generated protein embeddings, we assessed DHR by comparing it to methods on the standard SCOPe (structural classification of proteins) dataset37,38. SCOPe is a database of protein structural interactions that include the carefully curated ordering of domains from most known proteins in a hierarchy based on structural and evolutionary ties. We conducted studies to investigate how existing homolog retrieval methods and DHR aligned with these curated domain labels. While based solely on sequences, DHR can be compared to other previous sequence-level matching homolog extraction methods such as PSI-BLAST39, MMseqs2 (ref. 12) and DIAMOND40 (configurations are detailed in the Methods). We also included a profile-based method, HMMER41, and a structure-based method, Foldseek42, to better illustrate DHR’s effectiveness. Because the SCOPe dataset groups sequences into families on the basis of structural similarities, the sensitivity for SCOPe families reveals how much structural information these homolog extraction methods will capture. To begin, we used a representative sequence from each family as the query when searching the entirety of SCOPe. To locate all homologs belonging to the same family, we searched the database using representative sequences of each family. Hits (true positives, TPs) are sequences in the retrieved sequences that belong to the same family as the query. False positives (FPs) are defined as hits that are not within the family. Thus, a general comparison can be made between our approach and the baseline methods using their sensitivity rate (\(\frac{\rm{TP}}{\rm{TP}+{FN}}\), where FN is the number of false negatives; Methods).
To ensure a fair and comprehensive comparison, we limited our analysis to the top 200 sequences retrieved by each method. DHR demonstrated exceptional performance, detecting notably more homologs (with a sensitivity of 93%) than any other method tested. Another widely used and sensitive method, HMMER, achieved the second-best result with a sensitivity of 91%. In contrast, Foldseek, despite relying solely on structural information, performed similarly to the sequence-based PSI-BLAST (both at ~88% sensitivity). This finding suggests that sequence-based methods, including DHR, are capable of incorporating rich structural information. When using MMseqs2 as a baseline for all sequences and categorizing cases with MMseqs2 sensitivity below 0.34 as hard cases for further investigation, DHR still showed a high average sensitivity over 80% with a perfect sensitivity of 100% in many cases (Fig. 2b). We show that, on several sequences among these hard cases (Fig. 2b), such as d1w0ha, only DHR was able to identify the proper homologs while other methods could not (Fig. 2b and Supplementary Fig. 1). In addition to being ahead in terms of the average results, DHR outperformed the baseline methods in sensitivity for most SCOPe families (Fig. 2c). Moreover, we observed that all methods struggled as SCOPe family sizes increased while DHR maintained a more robust performance (Fig. 2e).
All methods used a single sequence as input unless especially mentioned. a, A t-distributed stochastic neighbor embedding visualization of the seven largest SCOPe family embeddings from DHR. b, Violin plot of recall (sensitivity) rate comparison of DHR to other methods on the SCOPe dataset of 5,065 queries. Right, the detailed performance of these methods within the hard cases (MMseqs2 recall rate below 0.34, colored in red). c, Head-to-head comparison of sensitivity between DHR and other methods. d, AUC-1FP metric comparison of different methods on the SCOPe family level (n = 5,065 queries) and the superfamily level (n = 2,065 queries). Data are presented as bar plots where the height of each bar represents the mean and error bars indicate the s.d. e, Regression plot of the recall rate against the SCOPe family size. f, Scatter plot of AUC-1FP against the number of sequence hits with the SCOPe dataset on UniRef90 (training set). Further visualization of the AUC-1FP of the zero-hit samples (n = 990) is provided as the mean value, with error bars indicating the s.d. g, The r.m.s.d. and TM-score relationship between the structure retrieved by DHR and the query structure according to the rank. We averaged the score on each rank with a central curve that connects the mean values of the data points, with two additional curves surrounding the central curve representing the s.d. A higher TM-score or lower r.m.s.d. indicates better structural similarity. h, Failure of k-mer-based methods on the c.55.3.5 family. The predicted structure similarity using DHR is much higher than that using k-mer-based methods, indicating that sequence-based methods using k-mers do not consider structural information. i, Two samples ranked highly by DHR for the c.55.3.5 family. SCOPe did not include them in the family but SCOP2 did. More potential problematic classification examples are shown in Supplementary Figs. 1–3 and Supplementary Table 2.
To strengthen our results, we also incorporated another standard metric, the area under the curve before the first FP (AUC-1FP; Methods), on the SCOPe dataset, which is independent of the sequences retrieved by DHR. Both DHR and HMMER showcased the best results, achieving scores of 89% and 88%, respectively (Fig. 2d). Interestingly, the performance of PSI-BLAST and Foldseek was also comparable to that of DHR, a deviation from what we observed with the sensitivity metric. Meanwhile, MMseqs2 showed a slight improvement, reaching 82%, yet still trailing behind DHR by approximately 7%. Although different metrics may yield slightly varied performances, DHR consistently maintained a high level of performance. Furthermore, when considering execution time, DHR greatly outperformed its counterparts. Despite Foldseek, PSI-BLAST and HMMER showing comparable results in terms of AUC-1FP, their execution times were markedly longer. DHR was twice as fast as Foldseek, over 20 times faster than PSI-BLAST and substantially quicker than the profile-based method HMMER, which requires a time-consuming process to build profiles (Supplementary Table 1). On the other hand, MMseqs2, while comparable to DHR in terms of execution time when limited to two iterations, greatly lagged in performance (Fig. 2b,d and Supplementary Table 1). Importantly, the superiority of DHR was not merely a result of replicating our training and searching data. When using BLAST to explore the SCOPe database against our training set UniRef90, we found that most samples yielded fewer than 100 hits (Fig. 2f and Supplementary Table 3). Interestingly, approximately 500 samples returned no hits at all, indicating that they are ‘unseen’ structures with respect to our dataset. While these structures might pose challenges to DHR, they did not impact other alignment-based methods that are not based on deep learning. Nevertheless, DHR continued to produce high-quality predictions, achieving an AUC-1FP score of 89% (Fig. 2f). This highlights DHR’s robustness and efficiency, proving its ability to handle data to which it has not been previously exposed. When extending our analysis to the superfamily level with more challenging remote homologs, all methods (DHR, HMMER, Foldseek and PSI-BLAST) experienced a notable performance decline, with an approximately 10% decrease across the board (Fig. 2d). Despite this drop, DHR managed to maintain a leading performance, achieving an AUC-1FP score of 80%, with HMMER, Foldseek and PSI-BLAST showing slightly lower results (Fig. 2d). We did not include HHblits43, another mainstream method for homology searching, in previous comparisons because it requires a database of hidden Markov models (HMMs) provided by HH-suite. We additionally benchmarked HHblits on its curated SCOP95 dataset intersected with the SCOPe superfamily dataset and DHR still achieved highly accurate performance and faster computing speed on this dataset (Supplementary Fig. 8).
During homolog retrieval, it was shown that DHR sequence embeddings include massive structural information (Fig. 2a) and that DHR’s accuracy on these retrieved homologs even surpasses that of structure-based alignment methods. This intriguing finding prompted us to explore further and revealed a correlation between DHR’s sequence similarity rankings and structural similarity. Structural similarity between two distinct proteins is computed by DeepAlign44, a protein structure analysis toolkit. DeepAlign superposes two protein structures and calculates the TM-score45 according to a specified residue mapping. Studies based on the average across all SCOPe samples suggested that the TM-score decreases and r.m.s.d. increases monotonously along with DHR rank, determined by the embedding similarity between queries (Fig. 2g). A concrete example on query d1w0ha shows the reliability of DHR and the potential cause of failure of k-mer-based methods (PSI-BLAST and MMseqs2; Supplementary Fig. 1). Because PSI-BLAST and MMseqs2 did not yield any hits for d1w0ha, we looked at the top five retrieved homologs of DHR, which all belonged to the same family as d1w0ha in SCOPe. When we use PSI-BLAST to align the mapping, the structure similarity only yielded about 0.3 on these top five homologs compared to the structure alignment (Fig. 2h). As a result, the top five homologs that achieved higher scores were classified into the same family by DHR yet undetected by PSI-BLAST. This implies that DHR can capture more structural information (>0.5) than sequence-similarity-based methods (<0.5)46, providing further evidence of its effectiveness in homolog retrieval.
Lastly, it is interesting to observe that DHR can offer structural information beyond the SCOPe families. We discovered two homologs scoring highly in DHR that mismatched with SCOPe labels, which were ranked third and fourth among the top five (Fig. 2i). When we look at the homologs retrieved from query d1w0ha, the TM-align score suggests that their similarity was strong enough to be classified into the same family and they were indeed clustered with the query in the ‘δ-endotoxin C-terminal domain’ family in the updated SCOP2 dataset47 (Supplementary Table 2). This suggests a potential misclassification in SCOPe and that DHR-learned embeddings could contain supplementary and diverse structural information. We also performed a thorough scan on all 10,000 SCOPe families and calculated the TM-score of the top 20 target structures retrieved by DHR against the query target structures. The results showed several cases of inconsistent classification at the superfamily level between SCOPe and other structure classification datasets (Supplementary Table 2).
Rapid construction of rich and diverse MSAs
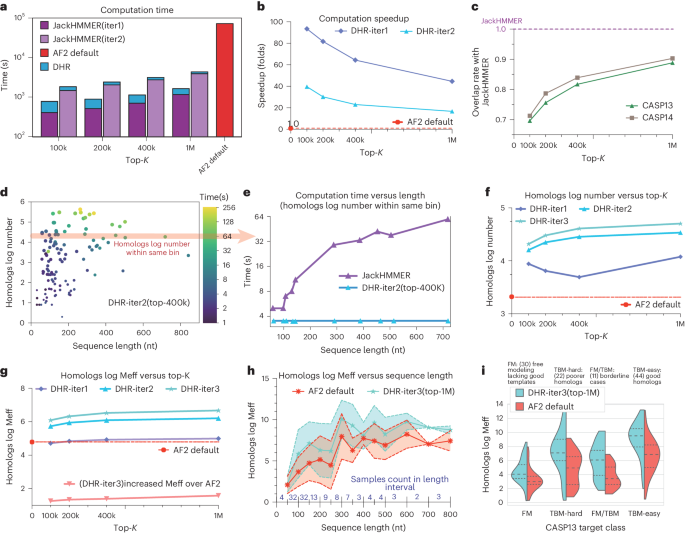
As a direct and critical downstream application of retrieving homologs, we could create an MSA with JackHMMER from homologs given by DHR and then compare it to the conventional AF2 default pipeline. Here, DHR served as a prefiltering method while JackHMMER used its multiple ungapped segment Viterbi (MSV) as a filtering method. This allowed us to assess both the quantity and the quality of MSAs constructed by DHR + JackHMMER in comparison to AF2 default (vanilla JackHMMER). Here, all MSA constructions were conducted using the same UniRef90 dataset. To accurately evaluate our performance, we also took into account different DHR or JackHMMER configurations, including DHR top {100k, 200k, 400k, 1M} and JackHMMER iteration {1, 2, 3}. We also included a DHR version without contrastive learning, ‘DHR-w/o con’, to demonstrate the necessity of our learning module. As JackHMMER’s search time is mainly dedicated to scanning and aligning, by leveraging the alignment-free nature of DHR, this process can be greatly expedited for a remarkable increase in overall efficiency. All configurations of DHR + JackHMMER ran faster than vanilla JackHMMER, AF2 default, on average (Fig. 3a). Our projections on UniRef90 at a 70M rate brought a 93-fold acceleration in the lightest configuration (DHR-iter1 (100k)) on average (Fig. 3b). We anticipate even greater improvements if our proposed strategy is applied to broader sequence pools.
a, Time expense at each step. Note that the y axis is set to a logarithmic scale. b, Overall improvement in speed of our method under different sensitivity settings compared to JackHMMER. The x axis controls the number of sequences retrieved by DHR and different colors denote the searching iterations using the subsequent JackHMMER. In the fastest setting, we achieved a 93-fold acceleration. c, Overlap ratio between our retrieved sequences and JackHMMER results. We conducted experiments on the CASP13DM and CASP14DM datasets with the highest overlap ratio above 90%, clearly demonstrating that our method builds MSAs rather similar to JackHMMER. d. Time consumption of searching two iterations with JackHMMER on CASP13DM samples relative to query sequence length and final homolog number. e, Selected samples (red arrow in d) with APPROXIMATELY 10,000 homologs in the MSA showing the effect of sequence length on searching time consumption. f, Logarithmic number of sequences in MSAs using DHR under different settings. The default MSAs built by the AF2 pipeline served as the baseline. g, The log Meff in the MSAs produced by DHR under different settings. h, The log Meff comparison for different sequence lengths using a central curve that connects the mean values of the data points, with two additional curves surrounding the central curve representing the s.d. i, Comparison of the Meff values of DHR and default AF2 in different CASP13 classes. Samples included queries with a good template (right) or with a poor or no template (left).
Surprisingly, despite being trained on a notably smaller dataset and using HHblits, DHR demonstrated an approximately 80% overlap with vanilla JackHMMER in constructing MSAs on UniRef90. This suggests that many MSA-related downstream tasks can be performed using DHR, yielding similar outcomes but with greater speed. We conducted this study on two test datasets, CASP13 and CASP14 domain sequences48. To ensure accuracy and reliability, our method only considers sequences with at least ten matching MSA homologs as valid and eliminates the remaining sequences. When using the top-1M configuration, DHR found around 90% of the same sequences as AF2 default JackHMMER at a greater speed. On the other hand, without contrastive learning and relying solely on ESM protein embedding, ‘DHR-w/o con’ achieved an overlap rate of only ~35% (Fig. 3c). As the quality of the MSAs is not directly associated with its overlap rate with JackHMMER, we continued to evaluate the quality of its MSAs using subsequent structure prediction applications. We discovered that DHR, when configured with 1M configurations and having the highest overlap rate, not only aligned closely with JackHMMER but also produced a variety of distinct MSAs, yielding the best outcomes (Fig. 6).
Not only did DHR obtain impressive computational acceleration and considerable overlapping rates but it was also capable of delivering increased homologs and MSAs for a query when compared to AF2 default (Fig. 3f). Because more and diverse MSAs could lead to a potential improvement of protein structure prediction1, such an increase is desired in MSA construction. Depending on the configuration, various levels of growth can be achieved. Among them, DHR-iter (1M) would experience the highest increase and a larger DHR top-K with more JackHMMER iterations would build more homologs. Furthermore, acquiring coevolutionary data from shorter sequences is more challenging because of fewer homologs in the database, which leads to a lower number of homologs collected with DHR (Fig. 3d). Nevertheless, another benefit of DHR is that it constructs the same number of homologs with varying lengths in a constant time, whereas JackHMMER scales linearly (Fig. 3e). Given that homologs or MSA may include redundant information, we used Meff introduced by Morcos et al.49 to further provide preliminary data where we believed that more nonredundant MSAs may lead to better quality and diversity. Our research indicated that DHR-iter1 and AF2 default produced a similar Meff, while the highest Meff was yielded by DHR-iter3, superior to the performance of AF2 default by a margin up to 1.5-fold (Fig. 3g). While fewer homologs were retrieved for shorter sequences, a similar trend could be also observed in MSA log Meff; for sequences under 50 amino acids, their computed MSA log Meff values were below 2, while the maximum log Meff was achieved on sequences between 250 and 300 amino acids (Fig. 3h). We also assessed DHR using several hand-crafted categories from CASP13 and CASP14 (Fig. 3i). The results were unequivocal; all categories experienced substantial benefits from using DHR, with notably greater increases in TBM-hard cases (hard-to-find from template-based modeling) than in other areas. This reinforces our conclusion that DHR is a promising approach for constructing MSAs across all categories.
DHR-constructed MSA boosts AF2-based structure prediction
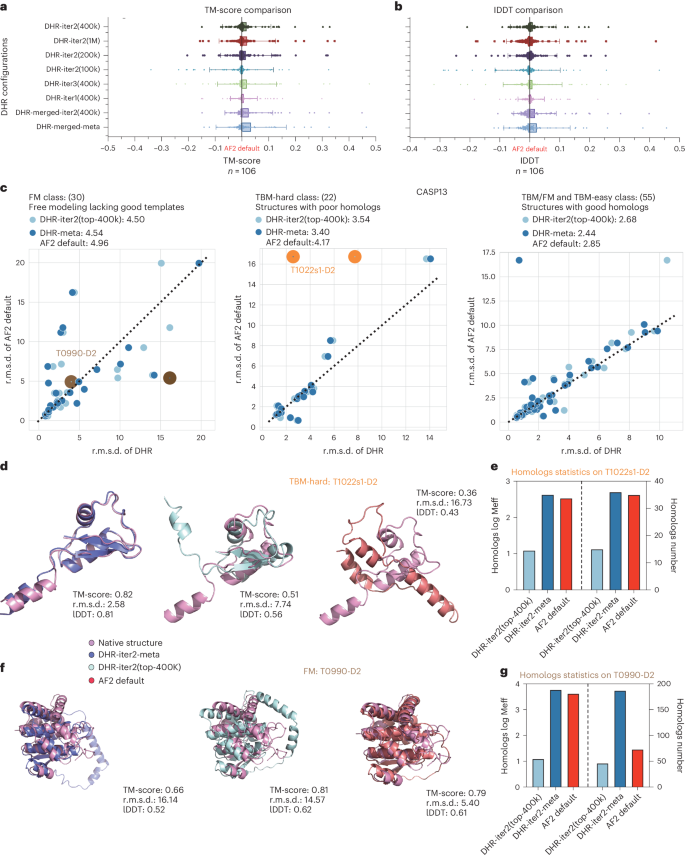
We previously revealed some statistical results of MSA log Meff to evaluate the quality of our MSAs built on retrieved homologs (Fig. 3g). Nevertheless, these evaluations mainly address diversity rather than quality. While there is no established standard for directly measuring MSA quality, we can indirectly estimate it by examining associated downstream applications, such as protein structure prediction. Here, we compared our results to those obtained by vanilla JackHMMER for predicting 3D structures using AF2 on the CASP13DM dataset. Consistent CASP target dataset results are detailed in Supplementary Table 6. We skipped the template searching phase and solely used UniRef90 to construct MSAs to prevent the feature stacking oriented from many databases and other potential impacts. An input query was given to obtain the top-K sequences (100k–1M) filtered by DHR and MSAs were constructed on these top-K sequences. To evaluate the accuracy of AF2’s prediction, five models were produced and only that with the highest predicted local distance difference test (lDDT), which denotes confidence ranging from 0 to 100, was taken into consideration. To further assess their quality, we calculated the TM-score and global lDDT score50 between native structures and modeled the results to measure how well our retrieved MSAs could aid in reconstructing a 3D structure. Then, we carried out a thorough analysis comparing all DHR setups to AF2 default. Standalone DHR performed similarly to AF2 default (TM-score ±0.02) and top-1M usually performed the best among all iterations (Fig. 4a,b and Supplementary Tables 5 and 7). While DHR produced a diverse MSA, we investigated whether it could be a supplement to AF2 default MSA and further boost the prediction results. We discovered that the merged MSA resulted in a performance boost (TM-score ±0.03) for DHR-merged-iter2 (top-400k), which merges MSAs from AF2 default with DHR-iter2 (top-400k). Moreover, the best performance was achieved by merging all MSAs from different DHR settings with AF2 default (DHR-merged-meta; Fig. 4a,b). These results suggest that, when used alone, DHR can be a fast and accurate replacement for the AF2 default MSA pipeline. When used in combination with the AF2 default MSA pipeline, DHR’s improvement over vanilla JackHMMER on MSA log Meff offers AF2 default with diverse and useful ‘structural’ information that aids in the structure reconstruction process.
a,b, Bar plots where the height of each bar represents the mean and error bars indicate the s.d. Plots show the all-atom TM-score (a) and lDDT (b) evaluation of different DHR configurations through their downstream structure prediction result. The default AF2 pipeline is set as a baseline to show the gain in performance. c, The DHR-produced MSA in structure prediction compared to AF2 default MSA. We show three head-to-head comparisons of different classes using the r.m.s.d. as an evaluation metric. d, Case study on the outlier case T1022s1-D2 identified as good by DHR. We show that the DHR-built MSA can produce a better structure than the default and that, by merging them, we can get even better predictions. e., The log Meff of the three MSAs in d. A large number of sequences may lack a quality guarantee. f, Case study on the outlier case T0990-D2 identified as bad by DHR. The merged result has a chain lying in the wrong position compared to the default prediction, leading to overall failure. We distilled each MSA and merged them again to get the correct prediction. g, The overall relationship between Meff and TM-score. Each blue point corresponds to one configuration of DHR.
In addition, we analyzed the performance of standalone DHR and DHR-meta in comparison to AF2 default on several types of CASP13DM structures. Because all models displayed nearly perfect accuracy (<3-Å r.m.s.d.) on the TBM-easy class, we investigated the two remaining challenging classes. T1022s1-D2 demonstrated that DHR-meta is superior to DHR-iter2 (400k) and AF2 default by 5-Å and 14-Å r.m.s.d., respectively (Fig. 4d). As a consequence, DHR-meta showed the highest MSA log Meff; such an accurate prediction may have been because of its diversified MSA (Fig. 4e). This conclusion may not apply to all protein cases but it is generally supported by average statistical evidence. This investigation was undertaken by comparing the average MSA log Meff to the average TM-score generated by various DHR configurations and merged models for all CASP13 targets (Fig. 4g). The regression line indicated a monotonously increasing trend between these two variables. We also analyzed a potential failure case T0990-D2, in which DHR-meta folded a helix in the incorrect position, resulting in an 11-Å increase in r.m.s.d. compared to AF2 default (Fig. 4f). After careful examination, we observed that the DHR-retrieved MSA was not necessarily the reason for this result. We tested an MSA-encoding pipeline with an identical MSA and found notably better performance, which we then dubbed DHR-distill. DHR-distill achieved 0.03-Å lower r.m.s.d. and 0.01 higher TM-score compared to AF2 default, which we did not investigate in detail. We believe that the AF2-like and JackHMMER black-box effect was the cause of this difference and further research could be conducted to gain a better understanding of this observation. Our trials revealed that, when MSA is combined with customized protein structure prediction techniques, it can boost accuracy in models. This approach allows us to quickly identify MSAs and use them for precise protein construct prediction from sequences.
MSA is an essential component for accurate protein structure prediction
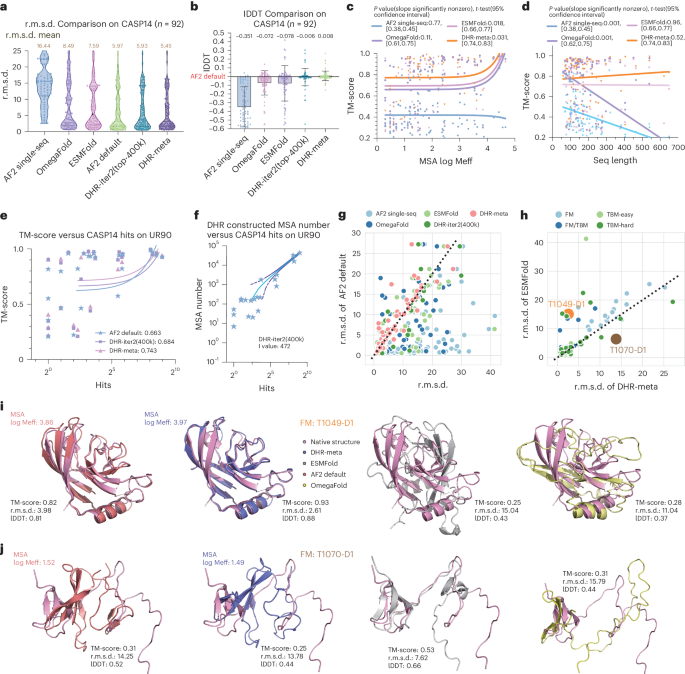
Recent advancements, such as RGN2 (ref. 36), ESMFold25 and OmegaFold32, use language models to bypass the process of producing MSAs. By doing so, they claim that MSAs are not necessarily needed for protein structures to be predicted successfully. These approaches also greatly reduce the processing time by eliminating the step of manually constructing MSAs. To examine the potential benefits that these language models could bring to protein structure prediction, we evaluated whether substituting MSA construction with language models on all CASP14DM targets would generate better results. To that end, we chose ESMFold and OmegaFold as our primary validation methods because they are open source with no data leakage related to any of the CASP datasets. AF2 Single-Seq was also included to conduct a model-based ablation study. They were compared to MSA-guided AF2 models, such as AF2 default, AF2-DHR-iter2 (400k) and DHR-meta. As anticipated, we achieved an r.m.s.d. improvement of 2.1 Å for DHR-meta over ESMFold and OmegaFold, where ESMFold performed slightly better. Moreover, the superior performance of DHR-meta did not arise from the AF2 default-constructed MSA as AF2-DHR-iter2 (400k) achieved a 0.03-Å lower r.m.s.d. compared to AF2 default (Fig. 5a,b and Supplementary Tables 6, 8 and 9).
a, Violin plot of r.m.s.d. comparison using different methods. b, Bar plot of lDDT score comparison, setting the AF2 result on CASP14 as the baseline, where the height of each bar represents the mean improvement of each method over AF2 default and error bars indicate the s.d. c, Relationship between MSA log Meff and TM-score on the predicted structures. d, Relationship between query sequence length and TM-score on the predicted structure. e, Relationship between the CASP14 sequence hits on UniRef90 (training data) and TM-score. f, Regression plot of the relationship between the CASP14 sequence hits on UniRef90 (training data) and DHR-constructed MSA log Meff, where the central curve shows the fitted mode and the two additional curves surrounding the central curve indicate the 95% intervals. g, Head-to-head comparison between AF2 default and other methods on CASP14 using r.m.s.d. as the evaluation metric. Each dot represents a sample, with the x axis showing the custom method result and the y axis showing the AF2 default distance. Dots above the reference line indicate that the method performed better than AF2 default. h, Head-to-head comparison between AF2 default and our method with merged MSAs on different classes using r.m.s.d. as the metric. Dots above the reference line indicate that our method achieved higher precision. Two outliers are shown in g,h. i, Case study of an outlier case T1049-D1 identified as good by DHR. j. Case study of an outlier case T1070-D1 identified as bad by DHR.
Furthermore, we investigated each case in CASP14DM to determine the causes for such disparity. In the more challenging cases where only a few MSAs were detectable, DHR-meta was observed to surpass language model-based methods (Fig. 5g), indicating that MSAs created by DHR can provide more critical structural information for 3D modeling. On simple cases with a large number of available MSAs, a language model could convey as much information as MSAs, resulting in the same performance (Fig. 5c). Examining yet another element impacting the reconstruction standard (sequence length), DHR-meta benefited from a slight TM-score boost as the sequence size increased. On the other hand, ESMFold remained stable while OmegaFold suffered from dramatic declines (Fig. 5d). We also noticed that algorithms such as OmegaFold exhaust the 32-GB memory of V100 GPUs, preventing them from completing the prediction pipeline for sequences longer than 650 amino acids, whereas our algorithm processed the sequence accurately. This indicates that a DHR-constructed MSA is indeed essential for high-resolution models, especially in cases with limited training data or long sequences.
To strengthen our claim, we conducted similarity tests using BLAST to search CASP14 targets against UniRef90 (our search database) and analyzed the number of hits. We observed that, on average, DHR achieved a higher TM-score than AF2 default and all methods showed increasing performance with more hits in the search database (Fig. 5e). For CASP14 samples that either had no hits or very few hits against UniRef90, DHR still managed to generate effective MSAs that enhanced structure prediction (Fig. 5e,f and Supplementary Tables 10 and 11). This resulted in an approximate increase of 0.01 in TM-score for CASP14 samples with zero hits. Furthermore, a detailed head-to-head comparison between DHR-meta and the most efficient single-sequence approach, ESMFold, was conducted for all CASP14 targets. Our analysis showed that DHR-meta outperformed ESMFold in nearly every case, especially in the most challenging category (free modeling (FM); no hit targets) (Fig. 5h). In particular, we demonstrate a specific sample where DHR-meta could identify more MSAs for such a challenging class, thus resulting in a more accurate prediction (Fig. 5i and Supplementary Table 11). Despite the few exceptions where DHR-meta lagged behind ESMFold, our study revealed that, in cases without any MSA, DHR-meta and AF2 default would underperform to the level of AF2 Single-Seq (Fig. 5j). Overall, our results demonstrated that MSA-based models can greatly improve prediction accuracy and efficacy when compared to language model-based approaches. We show that the use of MSAs is beneficial and should not be overlooked in protein structure prediction pipelines. Furthermore, we highlight the greater performance potential of our method in challenging cases with limited MSA data.
Scalability in large datasets and high-quality MSA generation
DHR proved its efficacy and ability in assisting structural prediction using the million-scale dataset UniRef90. We further present here that DHR is scalable to other datasets that are 100 times larger than UniRef90 and can still generate high-quality homologs or MSAs. Here, we performed our studies on BFM/MGnify, a combination of BFD and MGnify (2019_05), and included hundreds of millions of sequences in line with another standardized pipeline, ColabFold, which is a reproduction of AF2 using MMseqs2 to generate MSAs on BFM/MGnify by default. Specifically, we assessed the quality of DHR-generated MSAs through ColabFold on protein structure prediction against its default setting using CASP15 dataset. It is important to note that we did not retrain DHR on the large-scale dataset. Instead, the UniRef90-trained version was directly applied for MSA search inference. In contrast, within ColabFold, MMseqs2-BFD/MGnify was established with a profile database of BFD/MGnify, which is expected to offer natural advantages over DHR.
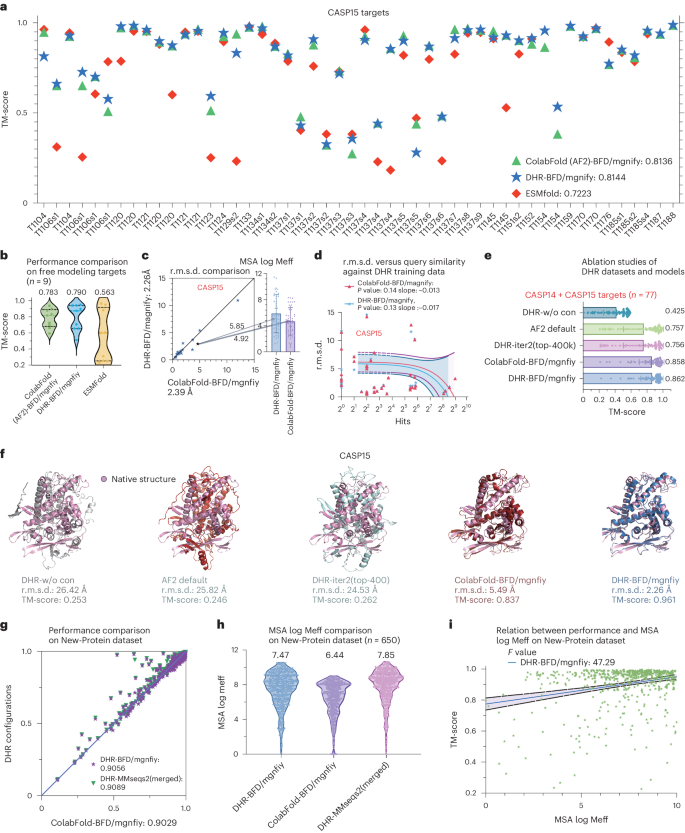
In evaluating the 50 CASP15 targets made available up to October 31, 2023 in terms of TM-score, DHR-ColabFold demonstrated a performance comparable to its default setting, MMseqs2, with a marginal improvement of approximately 0.01 (Fig. 6a, Supplementary Fig. 8 and Supplementary Table 9). Additionally, for most of the targets (36 of 50), DHR-ColabFold slightly outperformed ColabFold-MMseqs2. Despite this subtle enhancement, a review of the distinct MSAs generated by both methods (overlapping rate = 0.72) revealed varying rankings for each target. The observed variance suggests that DHR and existing homolog detection methodologies may serve similar roles. Notably, we included ESMFold, featured on the CASP15 leaderboard, to underscore the assertion that MSA is an indispensable component for precise structure prediction (Fig. 6a and Supplementary Table 12). While ESMFold showed an enhancement relative to its previous open version, it still lagged behind the MSA-based ColabFold (AF2) by approximately 0.09 in TM-score.
a, Comparative analysis of TM-score performance across 50 targets in CASP15. We used DHR-iter2 (top-400k) as the primary procedure for the MSA search, followed by ColabFold for downstream protein structure prediction. In contrast, ColabFold features its default pipeline using MMseqs2 for MSA search and ColabFold for structure prediction. Additionally, we included the results of ESMFold, an MSA-independent method based on single-sequence information, as extracted from the official results. b, Performance on nine FM targets in CASP15. c, Head-to-head r.m.s.d. comparison between ColabFold-MMseqs2 and ColabFold-DHR and their MSA log Meff results (represented as bar plots, where the height of each bar represents the mean and error bars indicate the s.d.). d, Regression plot of relationship between BLAST hits of a query (in CASP15) on our training set, UniRef90, against prediction r.m.s.d. values. More hits suggest a more similar query to our training set. The plots are presented as a central curve showing the fitted mode, with two additional curves surrounding the central curve showing the 95% intervals. e, Ablation studies of different DHR datasets and models on CASP14 + CASP15 (n = 77) targets, where DHR-iter2 (top-400k) was used. The results are presented as bar plots where the height of each bar represents the mean and error bars indicate the s.d. f. Visualization of a case study on CASP15 targets. This particular case was chosen on the basis of the notable increase in Meff observed when transitioning from the use of DHR-UniRef90 to DHR-BFD/MGnify. g–i, The results on our collected New-Protein dataset, which consists of 650 targets: head-to-head comparison of ColabFold-MMseqs2, ColabFold-DHR and DHR-MMseqs2 (merged) (g); MSA log Meff comparison (h); regression plot of the relationship between TM-score performance and MSA log Meff (i). The plots are presented as a central curve showing the fitted mode, with two additional curves surrounding the central curve showing the 95% intervals.
Predicting the structure of FM targets, especially those without structural templates in the Protein Data Bank (PDB) or with only limited MSAs, poses a great challenge in the field of structure prediction. In these complex scenarios, DHR stands out by generating meaningful MSAs, which improve structural predictions. DHR exceeded the performance of ColabFold, which uses MSAs constructed by MMseqs2, by 0.007 in terms of TM-score (Fig. 6b). Across all targets, DHR showed a slight increase in performance, evidenced by approximately 1.2-fold higher Meff compared to MMseqs2, translating to improved accuracy with a reduction of 0.15 Å in r.m.s.d. (Fig. 6c). Notably, for target T1129s2, DHR provided a 1.1 increase in Meff and an enhancement of 1.5 Å relative to ColabFold-MMseqs2 (Fig. 6c). Following the similarity tests in CASP14 and SCOPe, we also emphasize that the proficiency of DHR is not a consequence of simply memorizing query or hit pairs from its training data. We conducted a comprehensive similarity assessment of all targets against our training dataset with BLAST (Fig. 6d). Although it is anticipated that targets with greater similarity (larger hits) would enhance structure prediction, it is noteworthy that DHR’s performance remained relatively consistent even for the least similar targets (with zero hits), with an average r.m.s.d. of 4.8 Å (Fig. 6d).
While DHR scaled well on the large-scale BFD/MGnify datasets, we further show that DHR is ‘interpretable’, as a larger dataset improved its MSA quality, indicating that contrastive training is a necessary component within DHR (Fig. 6e). When benchmarked against different data scales, DHR combined with ColabFold (AF2) offered performance on par with (subtle improvement) JackHMMER or MMseqs2. The TM-score disparity across the two datasets reached up to 0.12 points, implying DHR’s scalability potential upon a subsequent increase in size of our training set. Notably, a DHR variant without contrastive training and relying solely on embedding similarity for homolog extraction had the most inferior performance (Fig. 6e). This underscores the importance of our designed learning mechanisms for optimal performance. Beyond statistical results, the interesting case of T1037-D1 actually showed that the variations between datasets and DHR models were sometimes large (Fig. 6f). When leveraging the UniRef90 dataset, DHR alongside AF2 default exhibited diminished efficacy, equating to MSAs constructed solely using protein embeddings, labeled ‘DHR-w/o con’. However, upon transitioning to the BFD/MGnify database, both DHR and MMseqs2 yielded notably higher MSA Meff scores, culminating in a 5.49-Å r.m.s.d. for ColabFold-MMseqs2 and an even lower r.m.s.d. for DHR accompanied by an increased extracted Meff (Fig. 6f and Supplementary Table 11).
While the CASP15 targets served as a blind test for DHR, the dataset comprised only 50 structures, which is relatively limited for extensive analysis. To address this, we introduced a new benchmark dataset, the New-Protein dataset, assembled from recently disclosed PDB structures with a cutoff date of January 1, 2022 to January 1, 2023, as well as a 70% similarity filter, ensuring that the 650 selected proteins were absent from our search and training set. On these targets, DHR was also able to generate more comprehensive and meaningful MSAs, achieving a log Meff approximately 1.0 higher than those created by MMseqs2 (Fig. 6h). Additionally, the integration of MSAs generated by DHR with those from JackHMMER in AF2 demonstrated similar effects; this integration approach was also successfully applied between DHR and MMseqs2, where their combined MSAs produced the highest log Meff and prediction accuracy across our dataset of 650 new protein targets, achieving a TM-score of 0.909 (Fig. 6g and Supplementary Table 13). Our statistical analysis on the New-Protein dataset confirmed that an increased MSA log Meff is indicative of better prediction performance (Fig. 6i). The above experiments demonstrated DHR’s scalability in constructing high-quality MSAs on large-scale datasets for normal proteins (CASP targets and PDB data), which include resolved protein structures. Additionally, we further investigated the challenges of disordered proteins, which are involved in various essential biological processes51. DHR exhibited robust capabilities by constructing MSAs for disordered proteins on a large-scale search database with high diversity (Supplementary Fig. 9).