Cell culture
All cells were cultured at 5% CO2 and 37 °C. Suspension cell lines including OCI-AML2, OCI-AML3, Nalm6, MOLM-13, Jurkat, Jeko1, SupT1, K562, HL-60 and Kasumi-1 were maintained by centrifuging cells, removing the medium and resuspending cells in fresh complete medium. Suspension cell cultures were split when cell density reached 2 million cells per ml. Culture medium used was 1× RPMI 1640 base medium (Thermo Fisher Scientific) with 1% penicillin–streptomycin (Pen/Strep) and 10% heat-inactivated FBS (Thermo Fisher Scientific). OCI-AML2 and OCI-AML3 cells were cultured in MEM α medium (Fisher Scientific) supplemented with 20% FBS (Fisher Scientific) and 1% Pen/Strep. A549, MC38, A673, SH-SY5Y, H4, U251, MDA-MB-231, B16F10 and primary mouse PDAC1-10 (ref. 56) cells were cultured in DMEM medium (Fisher Scientific) supplemented with 10% FBS (Fisher Scientific) and 1% Pen/Strep. LA-N-5 cells were cultured in DMEM (Fisher Scientific) supplemented with 20% FBS (Fisher Scientific) and 1% Pen/Strep. KNS-42 cells were cultured in DMEM (Fisher Scientific) supplemented with 10% FBS (Fisher Scientific), 1% sodium pyruvate and 1% Pen/Strep. UT-SSC-42B cells were cultured in DMEM (Fisher Scientific) supplemented with 10% FBS (Fisher Scientific), 1% NEAA and 1% Pen/Strep. Calu-1, HCT-116, MHH-ES-1 and U2OS cells were cultured in McCoy’s 5A medium (Fisher Scientific) supplemented with 10% FBS (Fisher Scientific) and 1% Pen/Strep. SaOS cells were cultured in McCoy’s 5A (Fisher Scientific) supplemented with 15% FBS (Fisher Scientific) and 1% Pen/Strep. HOS, MG-G3, FADU, DETROIT, HT1080 and HPAF-II cells were cultured in MEM medium (Fisher Scientific) supplemented with 10% FBS (Fisher Scientific) and 1% Pen/Strep. NCI-H520, KYSE-30, KYSE-140, SiMa, PFSK1, PC3, 22Rv1, ASPC1, BXPC3, SU86.86 and YAPC cells were cultured in RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% FBS (Fisher Scientific) and 1% Pen/Strep. OE21 and OE33 cells were cultured in RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% FBS (Fisher Scientific), 2% l-glutamine (Thermo Fisher Scientific) and 1% Pen/Strep. All the primary murine AML cells were cultured in X-VIVO 20 medium with gentamicin and PR1 (Lonza) supplemented with 5% FBS, recombinant murine IL-3 (10 ng ml−1, PeproTech), IL-6 (10 ng ml−1, PeproTech) and SCF (50 ng ml−1, PeproTech) and 1% penicillin–streptomycin–glutamine (Gibco). Cell lines were acquired from ATCC and the Sanger Institute Cancer Cell Collection unless otherwise noted. A full list of the cell lines’ source is provided in the Reporting summary. Cell cultures were periodically checked for Mycoplasma and maintained as Mycoplasma negative. Human cell lines employed were either not listed in the cross-contaminated or misidentified cell line database curated by the International Cell Line Authentication Committee or were previously verified by karyotyping.
Ex vivo culture of murine primary leukemia
Flt3ITD/+ (Flt3 internal tandem duplication) mice were kindly provided by G. Gilliland (Harvard Medical School, USA) and crossed with Rosa26Cas9/+ mice. Freshly isolated BM from 6–10-week-old female Flt3ITD/+;Rosa26Cas9/+ or moribund Npm1fl−cA/+;Flt3ITD/+;Rosa26Cas9/+, Npm1fl−cA/+;NrasG12D/+ mice were used. BM cells were exposed to erythrocyte lysis (BD Pharm Lyse, BD Biosciences), followed by magnetic bead selection of Lin− cells using the Lineage Cell Depletion Kit (Miltenyi Biotec) according to the manufacturer’s instructions. Lin− cells were cultured in X-VIVO 20 (Lonza) supplemented with 5% FBS (Life Technologies), 10 ng ml−1 IL-3 (PeproTech), 10 ng ml−1 IL-6 (PeproTech) and 50 ng ml−1 SCF (PeproTech) and 1% penicillin–streptomycin–glutamine. The retroviral constructs pMSCV-MLL-AF9-IRES-YFP and pMSCV-MLL-ENL-IRES-Neo were used with the package plasmid psi-Eco to produce retrovirus. Next, 293T cells (Life Technologies) were cultured and prepared for transduction in 10-cm plates. For virus production, 5 μg of the above plasmids and 5 μg of the psi-Eco packaging vector were transfected dropwise into the 293T cells using 47.5 μl TransIT-LT1 (Mirus) and 600 μl Opti-MEM (Invitrogen). The resulting viral supernatant was collected as previously described. Transduction of primary Flt3ITD/+;Rosa26Cas9/+ mouse cells was performed in six-well plates as mentioned above. After transduction, transduced cells were sorted for YFP (for MLL-AF9) or selected with neomycin (for MLL-ENL).
Dissection and culture of primary mouse prostate tissue
Normal prostate tissue was derived from Rosa26-LSL-Cas9 knockin mice on B6J (strain 26175, Jackson Laboratory), and Pten−/−;Trp53−/− prostate tumors were derived from a mouse model of prostate cancer of the same genetic background similar to Feng et al.57. Tissues were minced into small pieces and transferred into a gentleMACS C tube (Miltenyi Biotec) for enzymatic digestion using the Multi Tissue Dissociation Kit 1 (130-110-201, Miltenyi Biotec). Digestion was performed in a gentleMACS Dissociator using the program 37C_Multi_A. Following enzymatic dissociation, the cell suspension was pelleted, resuspended and filtered through a 70‐μm cell strainer. Normal and tumoral dissociated prostate cells were cultured as 3D organoids using the protocol and medium composition described by Drost et al.58. Cultures were maintained under these conditions for 6–7 days to enrich for epithelial cells, after which cells were maintained as monolayer cultures on plates coated with collagen I (354236, Corning) using the same medium composition.
Lentiviral vector production and infection
For virus production, 293FT cells were transfected with the lentiviral vector (lentiCRISPR-v2) either as an empty vector or containing NPM1 guide RNA together with the packaging plasmids psPAX2 (Addgene, 12260) and pMD2.G (Addgene, 12259). The viral supernatant was collected 48 and 72 h after transfection and concentrated overnight at 6,000g and 4 °C. A total of 1 × 106 cells and viral supernatant were mixed in 2 ml culture medium supplemented with 8 μg ml−1 polybrene (Millipore), followed by spinfection (60 min, 900g, 32 °C), and were further incubated overnight at 37 °C. The medium was refreshed on the following day, and the transduced cells were cultured further. Pellets for protein were collected 7 and 9 days after transduction.
Protein extraction and western blot for the knockout experiment
OCI-AML2 cells were transduced with either NPM1 gRNAs or control gRNAs. Cell pellets were collected on day 7 after transduction and lysed with whole-cell lysis buffer (0.2% Nonidet P-40, 50 mM Tris-HCl, pH 8.0, 450 mM NaCl, 1 mM EDTA), 1× protease inhibitor cocktail 1 (Merck), 1× phosphatase inhibitor 2 (Merck), 1× phosphatase inhibitor 3 (Merck) and 1 mM DTT (Epigentek)) and incubated on ice for 10 min. The lysates were then centrifuged at 20,000g and 4 °C for 10 min. The supernatant was transferred to a fresh tube and quantified using the Bradford assay (Bio-Rad). Following quantification, the samples were supplemented with 1× LDS sample buffer (Thermo Fisher Scientific) and 1× Sample Reducing Agent (Thermo Fisher Scientific), and they were then incubated at 70 °C for 10 min. Next, 10 μg protein was loaded. Western blotting was performed using SDS–PAGE gels, and samples were blotted onto a PVDF membrane. It was performed using the following antibodies: anti-NPM1 (FC8791), anti-H3 (Abcam, ab1220) as a loading control at a 1:1,000 dilution and goat anti-mouse IgG H&L, HRP conjugated (Abcam, ab205719) at a 1:10,000 dilution.
Blood counts
For blood counts, 20 μl blood was collected from the tail vein of mice using a capillary pipette containing anticoagulants (EDTA). The EDTA anticoagulated blood samples were used to obtain a complete blood count with a Vet ABC analyzer (Horiba ABX). Samples were counted no longer than 20 min after blood was drawn.
Real-time PCR
For Fig. 4a, genomic DNA was extracted from murine PB using the DNeasy Blood and Tissue Kit (Qiagen). Genomic DNA (10 ng) was used, and the levels of Cas9 and Gapdh were analyzed on a QuantStudio 5 real-time PCR instrument (Applied Biosystems) using PowerUp SYBR Green Master Mix (Applied Biosciences). The relative quantification of Cas9 was performed using the comparative cycle threshold (Ct) method against the housekeeping gene Gapdh. The primer sequences are listed in Supplementary Table 3.
PCR
For Extended Data Fig. 6b, genomic DNA was extracted from murine PB 2 weeks after injection using the DNeasy Blood and Tissue Kit (Qiagen). Primers for Flt3ITD were used for PCR amplification of genomic DNA (20 ng). The PCR product was analyzed by agarose gel electrophoresis, stained with GelRed. The 1 kb plus DNA Ladder (NEB) was used.
Antibody staining and flow cytometry analysis of AML cells
For primary murine AML experiments related to Fig. 2b and Extended Data Figs. 2b and 3b, 6–10-week-old C57BL/6 male mice were injected with 106 primary murine AML cells by intravenous injection from the indicated animal AML models, as described above. Upon animal sickness, BM was isolated and lysed in 0.85% NH4Cl for 5 min. Primary antibodies, at a concentration of 0.5 μg per reaction, either anti-NPM1 (mAb2) or the IgG2a isotype control (Bio X Cell), were precomplexed with a 1:1,000 dilution of the secondary antibody goat anti-mouse IgG Alexa Fluor 488 (Abcam) for 30 min. For intracellular staining, cells were first permeabilized with 0.1% Triton X-100 (Sigma) for 10 min at room temperature and rinsed with 2% FBS in 1× PBS. For cell surface staining (‘live cell’), cells were processed directly. BM cells were blocked in 2% FBS in 1× PBS for 30 min on ice and then stained with the precomplexed mix of antibodies as stated above. Cells were washed once with 150 µl of 2% FBS in 1× PBS and resuspended in 2% FBS in 1× PBS containing 0.1 µg ml−1 DAPI (Sigma). For intracellular staining, cells were fixed with 4% formaldehyde followed by a wash with 2% FBS in 1× PBS. Flow cytometry analysis was performed using a CytoFLEX instrument (Beckman Coulter) and analyzed using FlowJo (version 10, BD).
For primary murine AML experiments related to Fig. 4d, 6–10-week-old C57BL/6 male mice were sublethally irradiated with a whole-body dose of 5.5 Gy and then injected with 106 primary murine AML cells by intravenous injection from the indicated animal AML model, as described above. On day 15 after transplantation, mice were treated i.p. with a single dose of either 5 mg kg−1 of the mouse IgG2a isotype control antibody or 5 mg kg−1 of anti-NPM1 (mAb2) antibody. On day 18 after transplantation, PB was isolated and lysed in 0.85% NH4Cl for 5 min. Flow cytometric analysis of YFP+ cells was performed as above.
For Fig. 2c, K562 cells were transduced with the lentiviral cDNA constructs pKLV-TY1-NPM1-PURO or pKLV-TY1-NPM1c-PURO (mutant NPM1) or an empty pKLV-TY1-PURO vector control. Transduced cells were selected with puromycin, and then live cells were stained with either a mouse anti-B23 NPM1 antibody (Merck) or a mouse anti-Ty1 antibody (Diagenode) for 45 min followed by a staining with a 1:1,000 dilution of the secondary antibody goat anti-mouse IgG Alexa Fluor 488 (Abcam) for 45 min. Cells were washed once with 2% FBS in 1× PBS and finally resuspended in 2% FBS in 1× PBS containing 0.1 µg ml−1 DAPI. Flow cytometry analysis was performed as above.
For Extended Data Fig. 2g, K562 and OCI-AML3 cells were incubated with human Fc block (BioLegend, 101319). Afterward, they were incubated with WGA conjugated to fluorescein (Vector Laboratories, FL-1021-5) at a concentration of 1:1,000. When colabeled with FC8791 or mAb2, antibodies were precomplexed with anti-mouse IgG Alexa Fluor 647 at a ratio of 2:1 before being incubated with cells. The final concentration of FC8791 and mAb2 was 5 µg ml−1, and the secondary antibody was used at 2.5 µg ml−1. Cells were washed after binding, subsequently stained with DAPI and then applied to slides using a Cytospin. Images were obtained on a Leica TCS SP8 microscope.
For AML PDX experiments related to Extended Data Fig. 6e,f, 6–10-week-old SCID-CB17 female mice (Charles River, strain 236) were injected with 106 patient-derived AML cells by intravenous injection. BM was isolated and lysed in 0.85% NH4Cl for 5 min. Primary antibodies, at a concentration of 0.5 μg per reaction, either anti-NPM1 (mAb2) or the IgG2a isotype control (Bio X Cell), were precomplexed with a 1:1,000 dilution of the secondary antibody goat anti-mouse IgG Alexa Fluor 488 (Abcam) for 30 min. BM cells were blocked in 2% FBS in 1× PBS for 30 min on ice and then stained with the precomplexed mix of antibodies as stated above. Cells were washed once with 150 µl of 2% FBS in 1× PBS and resuspended in 2% FBS in 1× PBS containing 0.1 µg ml−1 DAPI. Flow cytometry analysis was performed as above.
For primary murine experiments related to Fig. 5d, freshly isolated BM from male 20-week-old WT and preleukemic Npm1fl−cA/+ or moribund Npm1fl−cA/+;Flt3ITD/+ mice was used. BM cells were exposed to erythrocyte lysis (BD Pharm Lyse, BD Biosciences), followed by magnetic bead selection of Lin− cells using the Lineage Cell Depletion Kit (Miltenyi Biotec) according to the manufacturer’s instructions. Primary antibodies, at a concentration of 0.5 μg per reaction, either anti-NPM1 (mAb2) or the IgG2a isotype control (Bio X Cell), were precomplexed with a 1:1,000 dilution of the secondary antibody goat anti-mouse IgG Alexa Fluor 488 (Abcam) for 30 min. BM cells were blocked in 2% FBS in 1× PBS for 30 min on ice and then stained with the precomplexed mix of antibodies as stated above. Cells were washed once with 150 µl of 2% FBS in 1× PBS and resuspended in 2% FBS in 1× PBS containing 0.1 µg ml−1 DAPI. Flow cytometry analysis was performed as above.
For primary murine AML experiments related to Fig. 5a,b, 6–10-week-old C57BL/6 male mice were sublethally irradiated with a whole-body dose of 5.5 Gy and then injected with 106 primary murine AML cells by intravenous injection from the indicated animal AML model, as described above. Upon animal sickness on day 18 after transplantation, BM was isolated and lysed in 0.85% NH4Cl for 5 min. BM cells were resuspended in 10% DMSO in FBS and stored at −80 °C for further applications. BM cells were thawed and suspended in PBS supplemented with 2% FBS and stained with biotin anti-mouse Ly-6A/E (SCA1) (BioLegend), biotin anti-mouse CD127 (BioLegend, 135005), biotin anti-mouse CD3 (BioLegend, 100201), biotin anti-mouse TER-119/erythroid cells (BioLegend, 116203), biotin anti-mouse/human CD45R/B220 (BioLegend, 103203), BV605 anti-mouse GR1 (BioLegend, 108439), BV650 anti-mouse CD11b (BioLegend, 101239), PerCP/Cy5.5 anti-mouse CD16/CD32 (BioLegend, 101323), PE anti-mouse CD93 (BioLegend, 136503), APC anti-mouse CD48 (BioLegend, 103411), APC/Fire 750 anti-mouse CD117 (c-Kit) (BioLegend, 135139), Zombie aqua viability dye (BioLegend, 423101) and BV421 streptavidin (BioLegend, 405226). All the antibodies were used at a 1:400 dilution, apart from the viability dye, which was used at a 1:1,000 dilution. The samples were then stained with either anti-NPM1 (mAb2) antibody or the IgG2a isotype control (Bio X Cell, BE0085), precomplexed with anti-mouse IgG Alexa Fluor 594 (Abcam, ab150108) as a secondary antibody. FMO controls were included in the experiments to provide a measure of spillover in each channel. This allows for correct gating in each experimental sample. Flow cytometry analysis was performed using the Cytek Aurora spectral analyzer and analyzed using FlowJo (version 10, BD). Data in this section were plotted using GraphPad Prism (version 9).
Antibody staining and flow cytometry analysis of solid cancer models
Primary antibodies, at a concentration of 0.5 μg per reaction, either anti-NPM1 (mAb2) or the IgG2a isotype control (Bio X Cell), were precomplexed with a 1:1,000 dilution of the secondary antibody goat anti-mouse IgG Alexa Fluor 488 (Abcam) for 30 min. A total of 5 × 104 cells for each model used in Fig. 6 (Fig. 6a,b,e,h) were blocked in 2% FBS in 1× PBS for 30 min on ice and then stained with the precomplexed mix of antibodies as stated above. Cells were washed once with 150 µl of 2% FBS in 1× PBS and resuspended in 2% FBS in 1× PBS containing 0.1 µg ml−1 DAPI (Sigma). Flow cytometry analysis was performed using the CytoFLEX instrument (Beckman Coulter) and analyzed using FlowJo (version 10, BD).
In vivo treatment of normal, primary murine AML and PDX models
For experiments related to Extended Data Fig. 4d,e, 16–20-week-old C57BL/6 male mice were used, which were housed at Boston Children’s Hospital. All mouse procedures and protocols were approved by the Animal Care and Use Committee of Boston Children’s Hospital and followed all relevant guidelines and regulations. After euthanasia, PB, liver, spleen and BM were harvested.
For experiments related to Extended Data Fig. 5a,b, 6–10-week-old C57BL/6 male mice were given i.p. injections of either anti-NPM1 (mAb2) or the IgG2a isotype control (Bio X Cell, BE0085) antibody at the indicated doses, once per week for a total of 4 weeks (total of four treatments). Weights were recorded, and PB from the tail vein was collected at the indicated time points. Weight measurements of the indicated mouse organs were taken from all treated cohorts at the end of the study, on the 28th day after initiation of the relevant treatments.
For experiments related to Fig. 4d–f and Extended Data Fig. 5c,d, 6–10-week-old C57BL/6 male mice were sublethally irradiated with a whole-body dose of 5.5 Gy. On day 12 after irradiation, mice were given i.p. injections of either anti-NPM1 (mAb2) or the IgG2a isotype control (Bio X Cell, BE0085) antibody at the indicated doses, once per week for a total of 4 weeks (total of four treatments). Weights were recorded, and PB from the tail vein was collected at the indicated time points. Weight measurements of the indicated mouse organs were taken from all treated cohorts at the end of the study, on the 66th day after irradiation.
For primary murine Npm1c;Flt3ITD/+/Cas9 AML experiments related to Fig. 4a–c and Extended Data Fig. 6a, 6–10-week-old C57BL/6 male mice were sublethally irradiated with a whole-body dose of 5.5 Gy and then injected with 106 primary murine AML cells by intravenous injection from the indicated animal AML model, as described above. On day 14 after transplantation, mice were given i.p. injections of either anti-NPM1 (mAb2) or the IgG2a isotype control (Bio X Cell, BE0085) antibody at the indicated doses, once per week for a total of 4 weeks (total four of treatments). Weight measurements of leukemic spleens were taken from each animal after humane endpoints were reached.
For primary murine MLL-AF9/Flt3ITD/+/Cas9 AML experiments related to Fig. 4g,h, 6–10-week-old NSG male mice were injected with 106 primary murine AML cells by intravenous injection from the indicated animal AML model, as described above. On day 14 after transplantation, mice were given i.p. injections of either anti-NPM1 (mAb2) or the IgG2a isotype control (Bio X Cell, BE0085) antibody at the indicated doses, once per week for a total of 3 weeks (total of three treatments). Weight measurements of leukemic spleens were taken from each animal after humane endpoints were reached.
For xenotransplantation AML experiments related to Fig. 4i, 6–10-week-old SCID-CB17 (Charles River, strain 236) female mice were injected with 2 × 106 OCI-AML3 human AML cells by intravenous injection. On day 14 after transplantation, mice were given i.p. injections of either anti-NPM1 (mAb2) or the IgG2a isotype control (Bio X Cell, BE0085) antibody at the indicated doses, once per week for a total of 3 weeks (total of three treatments).
For AML PDX experiments related to Fig. 4j,k, 6–10-week-old SCID-CB17 (Charles River, strain 236) female mice were injected with 106 patient-derived AML cells by intravenous injection. On day 14 after transplantation, mice were given i.p. injections of either anti-NPM1 (mAb2) or the IgG2a isotype control (Bio X Cell, BE0085) antibody at the indicated doses, once per week for a total of 3 weeks (total of three treatments). PB from the tail vein was collected on day 24 after transplantation.
For primary murine MLL-AF9/Flt3ITD/+/Cas9 AML experiments related to Fig. 5c, 6–10-week-old C57BL/6 male mice were sublethally irradiated with a whole-body dose of 5.5 Gy and then injected with 106 primary murine AML cells from primary recipients. For the primary recipients (related to Fig. 4d,e and Extended Data Fig. 6c,d), 6–10-week-old C57BL/6 male mice were sublethally irradiated with a whole-body dose of 5.5 Gy and then injected with 106 primary murine AML cells by intravenous injection from the indicated animal AML model, as described above. On day 15 after transplantation, mice were treated i.p. with a single dose of either 5 mg kg−1 of mouse IgG2a isotype control antibody or 5 mg kg−1 of anti-NPM1 (mAb2) antibody. On day 18 after transplantation, BM was isolated and processed accordingly for secondary transplantation as indicated above. Moreover, on day 18 after transplantation, PB from the tail vein as well as weight measurements of spleens, lungs and livers were taken from each animal after humane endpoints were reached.
All mice used in the study were housed in specific pathogen-free conditions in the UBS animal facilities of the University of Cambridge. All cages were on a 12–12-h light–dark cycle (lights on, 07:30) in a temperature-controlled and humidity-controlled room. Room temperature was maintained at 72 ± 2 °F (22.2 ± 1.1 °C), and room humidity was maintained at 30–70%. The animals were culled when leukemia-associated symptoms occurred or humane endpoints were reached. All animal studies were carried out in accordance with the Animals (Scientific Procedures) Act 1986, UK and approved by the Ethics Committee at the University of Cambridge. Randomization and blinding were not applied. All data in this section were plotted using GraphPad Prism (version 9).
In vivo treatment of mouse solid tumor models
For primary prostate carcinoma experiments related to Fig. 6c,d, 8–10-week-old C57BL/6 male mice were injected with 106 primary murine prostate carcinoma cells by subcutaneous injection (1:1 ratio of Matrigel and cancer cells) from the indicated animal model, as described above. For colorectal carcinoma experiments related to Fig. 6f,g, 8–10-week-old C57BL/6 male mice were injected with 5 × 105 MC38 cells by subcutaneous injection (1:1 ratio of Matrigel and cancer cells). For melanoma experiments related to Fig. 6i, 8–10-week-old C57BL/6 male mice were injected with 5 × 105 B16F10 cells by subcutaneous injection (1:1 ratio of Matrigel and cancer cells). On days 5, 7 and 9 after transplantation, mice were given i.p. injections of either anti-NPM1 (mAb2) or the IgG2a isotype control (Bio X Cell, BE0085) antibody at the indicated doses (total of three treatments). Tumors were dissected when sizes were approaching or reached the humane end point limit (1.2 cm2) as per animal license: on day 17 after transplantation for prostate tumors, on day 13 after transplantation for colorectal carcinoma tumors and on day 11 after transplantation for melanoma. All tumor sizes were measured using a digital caliper (Jodsen). All solid tumor models used in the study were housed in specific pathogen-free conditions in the UBS animal facilities of the University of Cambridge,. All cages were on a 12–12-hour light–dark cycle (lights on, 07:30) in a temperature-controlled and humidity-controlled room. Room temperature was maintained at 72 ± 2 °F (22.2 ± 1.1 °C), and room humidity was maintained at 30–70%. The animals were culled when humane endpoints were reached. All animal studies were carried out in accordance with the Animals (Scientific Procedures) Act 1986, UK and approved by the Ethics Committee at the University of Cambridge. Randomization and blinding were not applied. All data in this section were plotted using GraphPad Prism (version 9).
Cell surface biotinylation, immunoprecipitation and western blotting
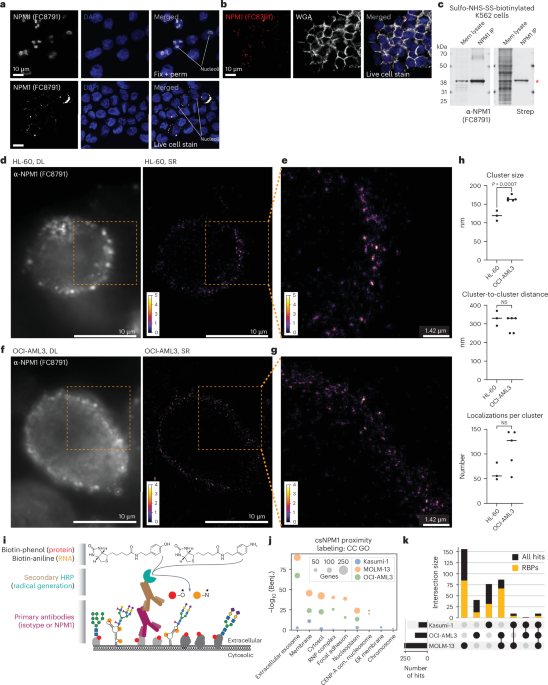
K562 cells were cultured as above. Cell surface protein labeling was accomplished using sulfo-NHS-SS-biotin (APExBIO) as described above, after which crude membrane fractions were isolated. To obtain cytosolic and membrane fractions24, suspension cells were directly resuspended in membrane isolation buffer (10 mM HEPES (Thermo Fisher Scientific), 250 mM sucrose (Sigma), 1 mM EDTA) at 5 million cells per 1 ml. Adherent cells were collected off the plate by scraping in ice-cold PBS, pelleted and then similarly resuspended at 5 million cells per 1 ml of membrane isolation buffer. Cells were rested on ice for up to 5 min, moved to a glass Dounce homogenizer (Sigma) and then homogenized using 40–80 strokes to obtain a resuspension of approximately 50% released nuclei. Overdouncing can cause nuclear rupture and contamination of the cytosolic fraction. After douncing, unbroken cells and nuclei were pelleted by centrifuging at 4 °C for 10 min at 700g. Supernatants (cytosol and membranes) were carefully transferred to a new tube, and pellets were discarded. The supernatants were again centrifuged at 4 °C for 30 min at 10,000g. Most (90%) of the supernatant was removed and saved as cytosolic fractions. The remaining supernatant was discarded, as it was near to the membrane pellet. The membrane pellet was gently washed with 500 µl of ice-cold 1× PBS (the pellet was not resuspended here), the tube was centrifuged briefly, and all supernatant was discarded to leave a cleaned membrane pellet. Finally, the membrane pellet was resuspended in 500 µl CLIP lysis buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl (Sigma), 1 mM EDTA, 10% glycerol (Thermo Fisher Scientific), 0.1% NP-40, 0.2% Triton X-100, 0.5% N-lauroylsarcosine). Both the cytosolic and membrane lysates were stored at −80 °C for later processing. After isolation and solubilization with CLIP lysis buffer, total protein quantification occurred using the BCA assay. For each sample, 5 μg protein was used for input, and 20 μg was used for either anti-NPM1 (FC8791)-coated bead protein A Dynabeads (Thermo Fisher Scientific) or streptavidin-coated bead (MyOne C1 beads, Thermo Fisher Scientific) enrichments. In both cases, 10 μl bead surrey was added to the membrane lysates in 100 μl of CLIP lysis buffer, and binding occurred at 4 °C for 16 h. After binding, the beads were washed three times with high-stringency buffer and then twice with 1× PBS. Proteins were released from the beads by heating at 85 °C for 10 min in 20 µl 1× LDS (Thermo Fisher Scientific) and 1 mM free biotin (Thermo Fisher Scientific). Input and enriched proteins were analyzed by Western blot, staining with anti-NPM1 antibody (Santa Cruz Biotechnology, sc-32256, FC8791) and streptavidin IR800 (LI-COR Biotechnology) and finally scanning on a LI-COR Odyssey CLx scanner. Fractionation as described above was performed without cell surface biotinylation for data in Fig. 1a, and western blots were developing using anti-NPM1 (Santa Cruz Biotechnology, sc-32256, FC8791), anti-NPM1c (Thermo Fisher Scientific, 32-5200), anti-β-actin (Santa Cruz Biotechnology, sc-47778) and anti-RPN1 (Santa Cruz Biotechnology, sc-48367) antibodies.
mAb2 design and synthesis
VH and VL sequences were grafted to the constant region of the mouse IgG2a heavy chain and the mouse λ light chain, respectively, to generate a new mouse antibody (mAb). These sequences were given to Curia Global. At Curia, the gene synthesis process involves overlapping oligonucleotide synthesis and assembly, followed by cloning into Curia’s proprietary high-expression mammalian vector. Production and mAb quality control were performed by Curia to produce a protein A-purified IgG fraction with low endotoxin (<1 EU per mg IgG) in 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4.
Live and fixed cell staining (human samples), flow cytometry and data analysis
Cells were cultured as described above and directly counted. Typically, 50,000 cells were used and blocked with Human TruStain FcX (Fc block, BioLegend) or Mouse TruStain FcX (Fc block, BioLegend) in flow cytometry buffer (0.5% BSA (Sigma) in 1× PBS) for at least 15 min on ice; cells were kept on ice from this point forward. For intracellular staining (‘fix–perm’), cells were first fixed with 3.7% formaldehyde for 10 min at 25 °C, rinsed once with 1× PBS and then permeabilized with 0.1% Triton X-100 (Sigma) for 10 min at 25 °C and finally rinsed once with 1× PBS. For cell surface staining (‘live cell’), cells were processed directly after live Fc blocking on ice. Precomplexed antibodies were added to live or fixed cells. To precomplex, primary unconjugated antibodies including mouse isotype (Santa Cruz Biotechnology, sc-2025) and anti-NPM1 (Santa Cruz Biotechnology, sc-32256, FC8791) and anti-Ty1 tag (Diagenode) were bound in solution (precomplexed) to a goat anti-mouse AF647 secondary antibody (Thermo Fisher Scientific, A32728) or a goat anti-mouse AF488 secondary antibody (Thermo Fisher Scientific, A28175) for at least 30 min on ice before use. The molar ratio was 2:1 (primary:secondary). To the blocked cells, precomplexed antibody was added at a final concentration of 1 µg ml−1 (primary antibody) and allowed to bind to cells for 60 min on ice. After staining, cells were centrifuged at 4 °C for 3 min at 400g, and the supernatant was discarded. All cell centrifugation steps took place using these conditions. Cells were washed once with 150 µl flow cytometry buffer, centrifuged under the same conditions and finally resuspended in flow cytometry buffer containing 0.1 µg ml−1 DAPI. Data collection occurred on a BD Biosciences LSRFortessa 3, and a gating strategy was used to isolate live single cells to examine antibody binding using FlowJo software (version 10, BD).
The frozen PB and BM samples obtained from healthy donors as well as patients with AML were processed with care to ensure as little cell lysis and high viability after thawing. Vials were warmed in a 37 °C water bath for 2–3 min and then completely thawed in 5 ml of ice-cold flow cytometry buffer. Cells were pelleted from this initial resuspension at 400g for 5 min at 4 °C. The supernatant was discarded, and cells were resuspended in 1 ml of fresh ice-cold flow cytometry buffer and counted. For each sample, 1 million cells per ml were taken for Fc blocking and staining according to the above protocol. Next, we stained the cells using the live protocol with the precomplexed isotype or anti-NPM1 (mAb2) antibodies for 30 min on ice in 100 µl flow cytometry buffer. After that, we centrifuged the cells and discarded the supernatant as before. The cells were then stained again with 200 ng of dye-conjugated cell type-specific antibodies (anti-human CD45 (HI30), 304024; anti-human CD34 (581), 343516; anti-human CD117 (104D2), 313204; anti-human CD33 (WM53), 303416; anti-human CD13 (WM15), 301710; anti-human HLA-DR (L243), 307604; anti-human CD3 (OKT3), 317308; and/or anti-human CD19 (HIB19), 302206; all BioLegend) on ice for 30 min in 100 µl flow cytometry buffer. Finally, the cells were pelleted again, the supernatant was discarded, one 200-µl flow cytometry buffer wash was performed, and the cells were finally resuspended in 200 µl flow cytometry buffer with 0.1 µg ml−1 DAPI for flow cytometry analysis. Healthy donor and AML patient samples were obtained with informed consent under (1) UK ethical approval (IRAS reference 340167, previously 149581, REC 07-MRE05-44). Additionally, healthy donor and AML patient samples were collected from patients located at the Dana-Farber Cancer Institute (DFCI) or Brigham and Women’s Hospital (USA). Samples were then processed and banked in the Pasquarello Tissue Bank in accordance with IRB-22-160 at the DFCI; we obtained assistance from the Hematologic Malignancies Data Repository to identify patient samples from the tissue bank that were bona fide AML. The HMDR also provided relevant information regarding disease characteristics, such as cytogenetics, mutations and immunophenotype. Samples were banked in accordance with DFCI Protocol 01-206: tissue and data collection for research studies in patients with hematologic malignancies, BM disorders and normal donors. Sample characteristics were obtained using DFCI IRB Protocol 22-160. This is a DFCI-specific tissue-banking protocol, which is not available publicly for review, but it can be shared upon request.
Confocal microscopy sample preparation, data acquisition and analysis
For suspension cells, culturing, counting and Fc blocking were carried out as described above. Samples for ‘live cell’ imaging were processed according to the live cell flow cytometry protocol noted above; however, after staining and washing, cells were fixed with 3.7% formaldehyde (37% stock, Sigma) for 30 min at 25 °C. Primary and secondary antibodies noted above were used but sequentially, rather than precomplexed: primary antibody was added at a final concentration of 2.5 µg ml−1 for 45 min on ice in flow cytometry buffer. After staining, cells were washed twice with 1× PBS and then stained with a secondary antibody at a final concentration of 2.5 µg ml−1. Secondary stains occurred for 30 min on ice and in the dark, after which cells were washed once with 1× PBS. A final fixation for ‘fix–perm’ samples was performed in parallel with the ‘live cell’ samples with 3.7% formaldehyde in 1× PBS for 30 min at 25 °C in the dark. Nuclei were stained with 0.1 µg ml−1 DAPI in flow cytometry buffer for 15 min at 25 °C. Suspension cells were applied to glass slides using a Cytospin centrifuge (Fisher Scientific): this was accomplished by centrifugation at 500g for 5 min in a Cytospin 1867. Finally, cells were mounted in ProLong Diamond Antifade Mountant (Thermo Fisher Scientific), and a coverglass was sealed over the sample with nail polish. All samples were then imaged on a Leica TCS SP8 STED ONE microscope. For all experiments, at least three regions of interest were acquired using a ×63 oil-immersion objective across one or more z slices. Leica’s line-sequential scanning method was used, and images were acquired at 1,024-by-1,024 resolution with a pinhole size of 1 AU. The DAPI channel was acquired with a PMT detector, while all other channels were imaged using hybrid detectors. For Extended Data Fig. 2i, the DAPI channel was imaged using a hybrid detector, and images were analyzed using ImageJ (version 1.54f). Next, using Imaris Microscopy Image Analysis software (Oxford Instruments), single slices of the confocal stack were analyzed.
Super-resolution imaging and reconstruction
Cells were prepared as described above for live cell imaging. Here, primary conjugated antibodies (AF647) were used directly for cell labeling and included mouse isotype (Santa Cruz Biotechnology, sc-24636-AF647) and anti-NPM1 (Santa Cruz Biotechnology, sc-32256-AF647, FC8791). To avoid cell movement during the SR acquisition, cells were immobilized on a glass-bottom plate precoated with poly-l-lysine (Sigma, P4707) and Cell-Tak (Corning, 354240). An overview of the method and processing can be found in Extended Data Fig. 3a. For single-molecule SR microscopy, we used direct stochastic optical reconstruction microscopy. To perform this, the PBS in which the cells were stored was replaced by a reducing oxygen scavenging buffer to induce blinking of fluorophores as described in the literature59. The blinking buffer consisted of 2 μl ml−1 catalase (Sigma), 10% (wt/vol) glucose (BD Biosciences), 100 mM Tris-HCl (Thermo Fisher Scientific), 560 μg ml−1 glucose oxidase (Sigma) and 20 mM cysteamine (Sigma). First, diffraction-limited images were obtained with low-intensity illumination of few W cm−2. Next, the laser power was increased to approximately 3 kW cm−2. Image acquisition was started after a short delay to ensure that most fluorophores were shelved into a dark state. The exposure time was 50 ms, and approximately 40,000 frames were recorded.
As the data were obtained with an sCMOS camera, which typically exhibit few pixels with deviating sensitivity (‘hot’ and ‘cold’ pixels), the obtained single-molecule data were corrected for individual pixels with abnormally high or low sensitivity first60. In total, 4,000 frames of a raw data stack were averaged. Hot and cold pixels, which are a systematic deviation, persist, in contrast to the random single-molecule signals. Each pixel was compared to its neighbors using 8-connectivity. If a deviation of more than 3% from the median of the neighboring pixel was observed, a correction factor that set the pixel to the median of its neighbors was recorded as previously described61. Otherwise, the pixel was not considered to be significantly brighter or darker. This yielded a correction mask, which was applied to all frames of the raw data. Finally, the no-light counts were subtracted from the pixel-corrected data.
For localization of single molecules, the Fiji plugin ThunderSTORM was used62. Each camera frame was filtered with a B-spline filter of order 3 and scale 2. Local maxima, corresponding to single-molecule signals, were detected with eight-neighborhood connectivity and a threshold of 1.1 or 1.2 times the standard deviation of the first wavelet level. Detected local maxima were fitted with a 2D Gaussian using least squares, and the position was recorded. Next, to account for single-molecule signals being active in multiple frames, merging of localizations was performed, using a maximum distance of 30 nm and a maximum of 5 off frames with no limit regarding on frames. Cross-correlation-based drift correction (magnification, 5; bin size, 5) was performed, followed by filtering of localizations (sigma of the point spread function between 60 and 270 nm, intensity below 37,800 photons, localization uncertainty smaller than 30 nm). For visualization, the final localizations were reconstructed as 2D histograms with a magnification of 5 (corresponding to a pixel size of 17.7 nm).
For cluster analysis, an automated pipeline was established using the raw list of localizations. First, Ripley’s H function was calculated on three areas with a large number of well-separated clusters. The resulting preferential cluster size from the three areas (which was, notably, always very similar) was averaged, multiplied by a correction factor of 0.45 and used as the seed radius for the following DBSCAN analysis. This DBSCAN script yielded all individual clusters, the total number of clusters, the average number of points per cluster and the spatial relation between clusters. The identified final clusters were then analyzed with respect to their spatial relation, size and number of localizations. This unbiased approach follows recommended analysis procedures recently described63. Crucially, each dataset was subjected to identical postprocessing and cluster analysis steps with no manual intervention, thus avoiding any biases arising from different parameter settings. Custom scripts used for this analysis can be shared upon request.
Cell surface proximity labeling of proteins, peptide generation and mass spectrometry data analysis
Samples were prepared similarly to the flow cytometry workflow as described above. However, rather than dye-conjugated secondaries, here an HRP-conjugated secondary (Thermo Fisher Scientific, 31430) was used. The isotype (control) or anti-NPM1 (FC8791) (target) primary unconjugated antibody was precomplexed with an appropriate secondary HRP antibody at 2:1 (primary:secondary) for at least 30 min on ice. Cells were grown as biological triplicate cultures, and typically 2.5 million cells were used per replicate per labeling experiment. Cells were collected from culture, washed of culture medium and resuspended on ice-cold flow cytometry buffer to which the Fc blocker was added for at least 15 min. After blocking, cells were adjusted to 1 million cells per ml of flow cytometry buffer, and then the precomplexed antibodies were added for staining at a final concentration of 2.5 µg ml−1. Staining occurred for 60 min at 4 °C on rotation, after which cells were pelleted, supernatants were discarded and cells were washed once with ice-cold 1× PBS. This wash is important to remove excess BSA in the flow cytometry buffer. Next, cells were gently but quickly resuspended in 980 µl of 100 µM biotin-phenol (Iris Biotech) in 1× PBS at 25 °C. To this, 10 µl of 100 mM H2O2 (Sigma-Aldrich) was quickly added, tubes were capped and inverted, and the reaction was allowed to proceed for 2 min at 25 °C. Precisely after 2 min, the samples were quenched by adding sodium azide and sodium ascorbate at a final concentration of 5 mM and 10 mM, respectively. Samples were inverted, and cells were pelleted at 4 °C. Samples were then washed sequentially once with ice-cold flow cytometry buffer and then twice with ice-cold 1× PBS, after which cell pellets were lysed in 500 µl CLIP lysis buffer, briefly sonicated to solubilize chromatin and frozen at −80 °C for later processing.
Once all the proximity labeling was complete, lysates were thawed in batches to perform the following steps in parallel. Total protein amounts were quantified using the BCA assay, and labeling efficiency and consistency were checked using western blotting. For biotin enrichment, we used the streptavidin western QC to determine the biotin signal across all samples first and then calculated the total µg of lysate that was needed from that sample to generate 5,000,000 units of streptavidin IR800 signal on the LI-COR system. This µg value was then used as the input mass for each of the replicates across all the proximity labeling samples for biotin capture and MS preparation. Samples were normalized to a final volume of 500 µl with CLIP lysis buffer, and, to each sample, 100 µl Pierce NeutrAvidin Agarose (Thermo Fisher Scientific) was added, and samples were incubated at 4 °C for 4 h on rotation. Beads were then washed twice with 1 ml of high-stringency buffer and twice with 1 ml of 4 M NaCl in 100 mM HEPES, all at 37 °C. Salts and detergents were then rinsed from the beads by sequentially washing twice with 1 ml 1× PBS, twice with 1 ml of LC–MS-grade water (Fisher Scientific) and finally with 1 ml of 50 mM ammonium bicarbonate. An on-bead trypsin digestion was then set up by adding 200 µl of 50 mM ammonium bicarbonate and 1 µg MS-grade trypsin to each sample and incubating for 16 h at 37 °C with occasional shaking. After digestion, the samples were acidified by adding formic acid at a final concentration of 0.5%. The solution containing released peptides was moved to a new tube, and the beads were rinsed twice with 300 µl LC–MS-grade water to capture any remaining peptides. All elution and wash samples for a given replicate were combined and reduced to a volume of <200 µl with a SpeedVac. Samples were then desalted using a C18 spin column (Thermo Fisher Scientific): preconditioned with 50% methanol in water and washed twice with 5% acetonitrile, 0.5% formic acid in water, the sample was bound twice to the column, and then the columns were washed four times with 0.5% formic acid in water. Finally, peptides were eluted into Protein LoBind tubes (Eppendorf) with two applications of 40 µl of 70% acetonitrile, 0.5% formic acid in water. Organic solvents were removed with a SpeedVac, and samples were fully dried with a lyophilizer. Resulting peptides were analyzed on the timsTOF Pro as described above.
For all peptides generated, we followed the same procedure for MS analysis and peptide database searching. Specifically, a nanoElute was attached in line to a timsTOF Pro equipped with a CaptiveSpray Source (Bruker). Chromatography was conducted at 40 °C through a 25-cm reversed-phase C18 column (PepSep) at a constant flow rate of 0.5 µl min−1. Mobile phase A was 98% water, 2% acetonitrile and 0.1% formic acid (vol/vol/vol), and phase B was acetonitrile with 0.1% formic acid (vol/vol). During a 108-min method, peptides were separated by a three-step linear gradient (5% to 30% B over 90 min, 30% to 35% B over 10 min, 35% to 95% B over 4 min), followed by a 4-min isocratic flush at 95% for 4 min before washing and a return to low organic conditions. Experiments were run as data-dependent acquisitions with ion mobility activated in PASEF mode. MS and MS/MS spectra were collected with m/z 100 to 1,700, and ions with z = +1 were excluded. Raw data files were searched using PEAKS Online Xpro 1.7 (Bioinformatics Solutions). The precursor mass error tolerance and fragment mass error tolerance were set to 20 ppm and 0.03, respectively. The trypsin digest mode was set to semi-specific, and missed cleavages were set to 3. The human Swiss-Prot reviewed (canonical) database version 2020_05 (downloaded from UniProt) and the common Repository of Adventitious Proteins (cRAP version 1.0, downloaded from the Global Proteome Machine Organization) totaling 20,487 entries were used. Carbamidomethylation was selected as a fixed modification. Oxidation (M), deamidation (NQ) and acetylation (N terminus) were selected as variable modifications. Raw data files and searched datasets are available on the Mass Spectrometry Interactive Virtual Environment (MassIVE), a full member of the ProteomeXchange Consortium, under the identifier MSV000092211. The complete searched datasets are also available in Supplementary Information.
To identify enriched proteins from these datasets, we took an approach that compared enrichment in the NPM1 to that of isotype antibodies. Results of the database search were first purged of non-human proteins and keratins as previously described. All proteins with fewer than two unique peptides were also filtered out of the list. Next, Excel was used to calculate the mean spectral count for each of the remaining proteins across triplicates. For each protein, the mean spectral counts associated with each antibody probe were divided by the mean spectral counts of the corresponding isotype, creating an enrichment factor of each protein in the proximity labeling over the isotype. Python (version 3.13) was then used to calculate the ratio of total protein isolated from proximity labeling with protein-targeting antibodies divided by that collected with isotype antibodies. Proteins from each set of proximity labeling data with enrichments either less than two or the previously described ratio were filtered from the dataset. The resultant lists comprise the hits associated with each round of proximity labeling. To perform GO term analysis, all proteins observed across all four fractions from each cell line were concatenated, creating a list of background proteins from each of the four cell lines tested. Lists of hits from the membrane hit pulldowns from each cell line were submitted to DAVID (https://david.ncifcrf.gov/) and run against their respective backgrounds. Next, for each category of GO term (BP, CC and MF), the union of the top four terms across all cell lines and their associated sizes and Benjamini values were plotted for comparison of enrichment across cell lines.
Anti-NPM1 molecular counting
OCI-AML3 cells were collected, washed with flow cytometry buffer and resuspended at 106 cells per ml. They were then blocked with Human TruStain FcX for 15 min with 5 µl of Fc block per million cells. They were washed again afterward. The cells were then partitioned for live cell and fixed cell staining. Cells planned for fixation were stained with 1 µl of fixable violet LIVE/DEAD stain per 106 cells per ml for 30 min. Afterward, cells were washed with flow cytometry buffer and resuspended in 4% PFA for 10 min at room temperature. They were washed with flow cytometry buffer and then resuspended in 0.1% Triton X for 5 min at room temperature. Again, the cells were washed with flow cytometry buffer. Live cells and fixed cells were then incubated with 500 ng FC8791–Alexa Fluor 647 in 100 µl for 30 min. Cells were washed twice afterward, and live cells were finally resuspended in DAPI. For molecular counting, Quantum Alexa Fluor 647 molecules of equivalent soluble fluorochrome beads were used from Bangs Labs. All samples were analyzed on the BD LSRFortessa. Standard curves were generated and fluorescence quantitation was performed as per Bang Labs’ QuickCal analysis tool.
Cell surface proximity labeling of RNAs and gel analysis
Samples were prepared here in a similar manner as described above for the proximity labeling of proteins with key differences. The HRP-conjugated secondary antibody was exchanged for protein A–HRP (Cell Signaling) and used at the same molar ratio: two primary antibodies per one protein A–HRP molecule. The biotinylation reagent used here was biotin-aniline (Iris Biotech); it was used at a final concentration of 200 µM in 1× PBS for the labeling reaction, and the labeling reaction was allowed to proceed for 3 min at 25 °C. After pelleting the cells from the quenching reaction, cells were directly lysed, and total RNA was isolated as described before25. Briefly, RNAzol RT (Molecular Research Center) was used to lyse cell pellets by placing the samples at 50 °C and shaking for 5 min. To phase separate the RNA, 0.4× volumes of water was added, and samples were vortexed, allowed to stand for 5 min at 25 °C and lastly centrifuged at 12,000g at 4 °C for 15 min. The aqueous phase was transferred to clean tubes, and 1.1× volumes of isopropanol was added. The RNA was then purified over a Zymo column (Zymo Research). For all column cleanups, we used the following protocol. First, 350 μl of pure water was added to each column, samples were centrifuged at 10,000g for 30 s, and the flowthrough was discarded. Next, precipitated RNA from the RNAzol RT extraction (or binding buffer precipitated RNA, below) was added to the columns, which were centrifuged at 10,000g for 10–20 s, and the flowthrough was discarded. This step was repeated until all the precipitated RNA was passed over the column once. Next, the column was washed three times in total: once using 400 μl RNA Prep Buffer (3 M guanidine hydrochloride (GuHCl) in 80% ethanol) and twice with 400 μl 80% ethanol. The first two centrifugation steps were at 10,000g for 20 s, and the last one was for 30 s. RNA was then treated with proteinase K (Ambion) on the column. Proteinase K was diluted 1:19 in water and added directly to the column matrix and then allowed to incubate on the column at 37 °C for 45 min. The column top was sealed with either a cap or Parafilm to prevent evaporation. After the digestion, the columns were brought to room temperature for 5 min; lowering the temperature is important before proceeding. Next, eluted RNA was centrifuged out into fresh tubes, and a second elution with water was performed. The mucinase StcE (1.5 μg for every 50 μl of RNA, Sigma-Aldrich) was added to the eluate, and samples were placed at 37 °C for 30 min to digest. The RNA was then cleaned up again using a Zymo column. Here, 2× RNA Binding Buffer (Zymo Research) was added, samples were vortexed for 10 s, and then 2× (samples + buffer) of 100% ethanol was added, and samples were vortexed for 10 s. The final RNA was quantified using a NanoDrop. In vitro RNase or sialidase digestions took place by digesting 50 μg total RNA with one of the following: nothing, 4 μl RNase Cocktail (Thermo Fisher Scientific) or 4 μl of α2-3,6,8,9-neuraminidase A (NEB) in 1× NEB Glyco Buffer 1 (NEB) for 60 min at 37 °C. After digestion, RNA was purified using a Zymo column as noted above and was then ready for gel analysis.
To visualize the labeled RNA, it was run on a denaturing agarose gel, transferred to a nitrocellulose membrane and stained with streptavidin25. After elution from the column as described above, the RNA was combined with 12 μl of Gel Loading Buffer II (95% formamide (Thermo Fisher Scientific), 18 mM EDTA (Thermo Fisher Scientific), 0.025% SDS) with a final concentration of 1× SYBR Gold (Thermo Fisher Scientific) and denatured at 55 °C for 10 min. It is important to not use Gel Loading Buffer II with dyes. Immediately after this incubation, the RNA was placed on ice for at least 2 min. The samples were then loaded into a 1% agarose, 0.75% formaldehyde, 1.5× MOPS Buffer (Lonza) denaturing gel. Precise and consistent pouring of these gels is critical to ensure a similar thickness of the gel for accurate transfer conditions; we aim for solidified gels approximately 1 cm thick. RNA was electrophoresed in 1× MOPS at 115 V for between 34 and 45 min, depending on the length of the gel. Subsequently, the RNA was visualized with a UV gel imager, and excess gel was cut away; leaving ~0.75 cm of gel around the outer edges of sample lanes will improve transfer accuracy. The RNA was transferred with 3 M NaCl, pH 1 (with HCl) to a nitrocellulose membrane for 90 min at 25 °C. After transfer, the membrane was rinsed with 1× PBS and dried on Whatman paper (GE Healthcare). Dried membranes were rehydrated in Intercept Protein-Free Blocking Buffer, TBS (LI-COR Biosciences) for 30 min at 25 °C. After blocking, the membranes were stained using streptavidin IR800 (LI-COR Biosciences) diluted 1:5,000 in Intercept blocking buffer for 30 min at 25 °C. Excess streptavidin IR800 was washed from the membranes using three washes with 0.1% Tween-20 (Sigma) in 1× PBS for 3 min each at 25 °C. The membranes were then briefly rinsed with PBS to remove the Tween-20 before scanning. Membranes were scanned on a LI-COR Odyssey CLx scanner (LI-COR Biosciences).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.