Synthesis of ISM012-042
The synthesis of ISM012-042 is laid out in detail in Supplementary Information 1.
In vitro experiments
Crystallization and data analysis
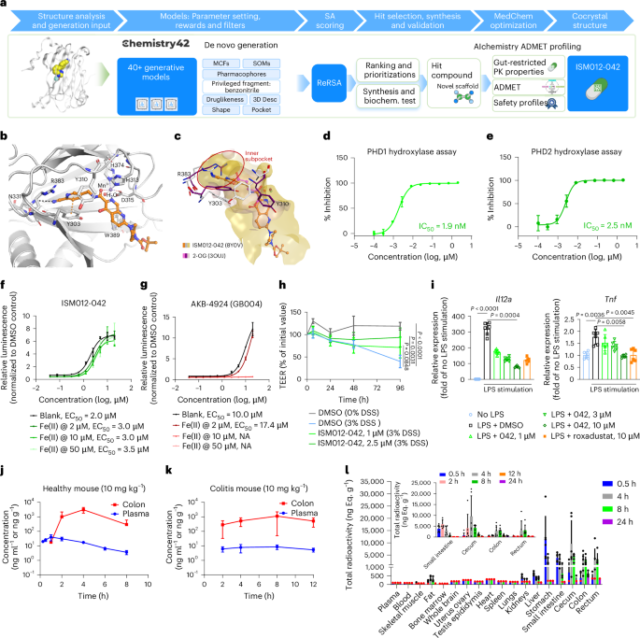
Recombinant PHD2181–426 protein was expressed in Escherichia coli and purified as previously described22. Protein sample was mixed with crystallization solution composed of 0.2 M NH4 phosphate monobasic, 0.1 M Tris pH 8.5 and 50% MPD using the sitting drop vapor diffusion method. The crystal structure of PHD2 (P181–E407) cocrystallized with MnCl2, ISM012-042 and 5% PEG smear appeared after 7 days at 20 °C. A crystal with the proper size was isolated and quickly cooled in liquid nitrogen for data collection. The crystals producing high-quality diffraction spots were chosen for full dataset collection at the Shanghai Synchrotron Radiation Facility synchrotron source. The diffraction data were indexed and integrated with XDS (version January 31, 2020) and scaled by aimless (CCP4 7.1). The structure was solved by molecular replacement with Phaser (CCP4 7.1). Refinement was carried out through multiple rounds with Coot (version 0.9.5) and refmac5 (CCP4 7.1). Crystallographic parameters and data collection statistics are described in Supplementary Table 10.
ADMET profiling
Metabolic stability in liver microsomes
ISM012-042 at 1.00 µM was incubated with CD1 mouse, SD rat, beagle dog, cynomolgus monkey and human liver microsomes with a reduced nicotinamide adenine dinucleotide phosphate (NADPH)-regenerating system in a 37.0 °C water bath for up to 60 min. The samples at 0, 5, 15, 30, 45 and 60 min were analyzed using liquid chromatography coupled with tandem mass spectrometry (LC–MS/MS; SCIEX API400 MS instrument coupled to a Waters ACQUITY ultrahigh-performance LC (UPLC) instrument). The residual percentage and the clearance rate of ISM012-042 were calculated from the measured concentration.
Permeability assay
Caco-2 cells at passage number 42 were seeded on 96-well transport inserts and cultured for 21 days before being used. ISM012-042 was dosed bidirectionally at 1.00 µM with or without 10.0 µM GF120918, a potent inhibitor of efflux transporters such as P-glycoprotein and breast cancer resistance protein. The plate was incubated for 2 h at 37.0 °C with 5.0% CO2 at saturated humidity without shaking. Samples were taken at 0 and 120 min after incubation and analyzed by LC–MS/MS. The apparent permeability (Papp, cm s−1) was calculated as follows, VR is the solution volume in the receiver chamber, CR is the final concentration of the test compound in the receiver chamber and C0 is the initial concentration of the test compound in the donor chamber:
$${P_{\rm{app}}}=\frac{\rm{VR}}{\rm{Area}\times {Time}}\times \frac{\left[{\rm{drug}}\right]{\rm{receiver}}}{\left[{\rm{drug}}\right]{\rm{initial},{donor}}}=\frac{\rm{VR}}{\rm{Area}\times {Time}}\times \frac{\rm{CR}}{C_0}$$
Cytochrome P450 inhibition
Pooled human liver microsomes were incubated in the presence of ISM012-042 (at 0, 0.000300, 0.00100, 0.00300, 0.0100, 0.0300, 0.100, 0.300 and 1.00 µM), NADPH and a selective substrate for each cytochrome P450 isoform (midazolam and testosterone) at 37 °C in a water bath. Reactions were terminated by adding 200 µl of stop solution. Formation of the selective metabolite from its substrate was determined by LC–MS/MS and reported as enzyme activities (pmol min−1 mg−1 protein) of each cytochrome P450 isoform. IC50 values were determined using a three-parameter or four-parameter logistic equation.
PK analysis of ISM012-042
All animal experiments were conducted by WuXi AppTec. All animals received a single oral dose after fasting overnight with free access to water and food was provided at 4 h after dosing. The blood samples were collected in K2 EDTA-coated tubes at 0.25, 0.5, 1, 2, 4, 6 and 8 h after dosing. Each group was killed by CO2 inhalation at 1, 2, 4 and 8 h after dosing and the colon tissue was collected. The dissected colon was washed with saline and homogenized by adding five volumes (w/v, 1:5) of precooled methanol and PBS (v/v, 1:2). The plasma and colon homogenate concentrations were determined by protein precipitation and LC–MS/MS detection (SCIEX Triple Quad 6500+ or 7500 MS instrument coupled to a Waters ACQUITY UPLC I-Class PLUS system). The PK parameters of plasma and colon were then calculated using noncompartmental analysis (Phoenix WinNonlin software, version 8.3.5, Certara). The protocol and any amendments or procedures involving the care or use of animals on this study were reviewed and approved (assurance identification numbers PK01-001-2019v1.0 and NJ-20220531-Rats) by the WuXi AppTec Institutional Animal Care and Use Committee (IACUC) before the initiation of such procedures.
Tissue distribution analysis of 14C-labeled ISM012-042
Sprague–Dawley (SD) rats were divided into seven groups (n = 3 per sex per time point) and administered a single oral dose of 14C-labeled ISM012-042 at 30 mg 100 μCi−1 kg−1. Four groups were anesthetized by isoflurane inhalation at 0.5, 4, 8 and 24 h after dosing. The heart, lung, liver, spleen, kidney, whole brain, small intestine, cecum, colon, rectum, stomach wall, skeletal muscle, body fat, bone marrow, reproductive organs (testis and epididymis for males; uterus and ovaries for females) and blood samples were collected. Three groups were anesthetized at 2, 12 and 48 h after dosing, after which the small intestine, cecum, colon and rectum were collected. A total of 12 SD rats (six males and six females) were administered a single oral dose of 14C-labeled ISM012-042 at 30 mg 100 µCi−1 kg−1. Urine, feces and cage wash up to 168 h after dosing were collected from six intact rats (three males and three females) for mass balance; bile, urine, feces and cage wash up to 72 h after dosing were collected from six bile-duct-cannulated rats (three males and three females) for biliary excretion. The TRA of plasma, bile, urine and cage wash samples was directly analyzed by liquid scintillation counting (LSC; PerkinElmer Instruments). Blood samples, fecal samples homogenized in 50% isopropanol in water (v/v), skeletal muscle, body fat, bone marrow and any other tissues were homogenized as needed and mixed with lytic agent SOLVABLE, heated in a water bath and analyzed by LSC. The blood samples were mixed with a hydrogen peroxide solution in water (7:93, v/v) before cocktail scintillation. Colon samples were homogenized on wet ice using homogenizing buffer (15 mmol L−1 phosphate buffer and methanol, 2:1, v/v) in a ratio of 1:5 (1 g of colon with 5 ml of buffer). The TRA values determined in bile, urine, feces and cage wash were used to calculate the recovery percentage of the administered dose.
AlphaScreen PHD hydroxylase assay
Compound screening assays were performed at Pharmaron. The final concentrations of the reaction for the respective PHD isoforms included recombinant PHD1 (Active Motif, 81064), PHD2 (Active Motif, 81065) or PHD3 (Active Motif, 81033), 2 μM 2-OG for PHD1 and PHD2 or 10 μM 2-OG for PHD3, biotinylated CODD peptide (Sangon; 3:1 ratio of enzyme to CODD peptide), l-ascorbic acid, 10 μM Fe(II) and inhibitors with 2% DMSO. Preincubated donor–acceptor bead mix (AlphaScreen streptavidin-conjugated donor and protein A-conjugated acceptor beads; PerkinElmer) with anti-HIF1α hydroxy-P546 antibody (Cell Signaling, 3434) was then added to the reaction mixture for 30 min at room temperature. The reaction was quenched with 5 μl of 30 mM EDTA. The luminescence signal was measured using an Envision (Perkin Elmer) plate reader. The data were analyzed using GraphPad Prism 8.0. For the examination of Fe(II)-dependent activities, the final Fe(II) concentration in the reaction was changed to the indicated level.
Enzymatic assay for FIH hydroxylase activity and JMJD2D, JMJD2E, JMJD1B and UTX demethylase activity
Protocols for the in vitro HTRF hydroxylase assay23 and the AlphaLISA histone demethylase assays24 were reported previously. For the FIH (abcam, ab86916) hydroxylase assay, the final Fe(II) reaction concentration was 5 μM. For the lysine demethylase assays, recombinant JMJD2D (Active Motif, 31459), JMJD2E (BPS Bioscience, 50118), JMJD1B (Active Motif, 31429) or UTX (Active Motif, 31460) was incubated with Fe(II) at 2 μM.
Cell lines
Caco-2 with HiBiT-tagged HIF1α was constructed by inserting the HiBiT coding sequence (Wuxi AppTec) at the C terminus of the endogenous HIF1A gene in Caco-2 cells (American Type Culture Collection, HTB-37) using clustered regularly interspaced short palindromic repeats and Cas9 and cultured in Eagle’s minimum essential medium containing 20% FBS and 1% penicillin–streptomycin at 37 °C with 5% CO2.
Mycoplasma testing
All cell lines were checked for Mycoplasma by WuXi AppTec and Shanghai Genechem.
Cell culture conditions
All cell lines were cultured at 37 °C in a humidified atmosphere of 95% air and 5% CO2.
Detection of HiBiT tag
Caco-2 HIF1α–HiBiT cells were seeded in a 384-well plate (5.5 × 103 cells per well for 48 h). The cells were treated with compound (10 μM, ten doses, in duplicate for ISM012-042 or GB004, freshly prepared before added to the medium) for 6 h. Cells were loaded with 0, 2, 10 or 50 μM Fe(II) sulfate in the medium for 20 min before DMSO or compound addition. HIF1α–HiBiT abundance in Caco-2 HIF1α–HiBiT cells was measured after 6 h through incubation with the Nano-Glo HiBiT lytic detection system and luminescence was measured using Envision. The HIF1α–HiBiT abundance in each well was normalized to the abundance of DMSO as a function of relative luminescence units (RLU). Abundance was measured in independent triplicates. After normalization, RLU values were used for regression analysis and EC50 curve fitting using GraphPad Prism.
TEER assay
Caco-2 cells were seeded at 5.0 × 104 cells per well within a 24-well transwell plate with 0.4-µm-pore polyester membrane inserts. The medium was replaced every 2 or 3 days. The TEER values of Caco-2 cell monolayers were monitored using a Millipore Millicell ERS-2 voltohmmeter once daily before monolayer establishment. When the TEER values reached a plateau (about 14 days after seeding) and were stable for the following 3–4 days, an efficient barrier function was considered established and ready for compound treatment. The monolayers were treated with ISM012-042 (1 or 2.5 μM) or DMSO in six replicates for 24 h before 3% DSS (w/v) treatment in both compartments of the transwell membrane for another 24 h along with the compound treatment. TEER values were obtained at 0, 6, 24, 48 and 96 h after the addition of DSS.
Immunoblotting
Confluent Caco-2 cells were incubated for 6 h in a medium supplemented with inhibitor, roxadustat positive control or DMSO. Then, 20 μM MG-132 (MedChemExpress, HY-13259) was added to Caco-2 cells for the last 4 h of a 6-h treatment with PHD inhibitors or DMSO. The hydroxy-HIF1α and total HIF1α proteins in the whole-cell lysates were assessed by western blotting and detected using anti-HIF1α (P564) (Cell Signaling Technology, 3434; 1:1,000) and anti-HIF1α (Cell Signaling Technology, 36169S; 1:1,000) antibodies with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a loading control (Merck, MAB374; 1:2,000). GAPDH band intensities were used to normalize total HIF1α or hydroxylated signals. The blots were visualized with an ImageQuant LAS 4000 camera (GE Healthcare).
Immunofluorescence cell staining of tight-junction protein
Briefly, Caco-2 cells were seeded on 14-mm-diameter coverslips precoated with 0.1% gelatin at 1 × 106 cells per well within a 12-well cell culture plate and pretreated with 3 μM ISM012-042 or DMSO for 1 h before tight-junction damage by 3% DSS (MP Biomedical, 216011050). After 24-h DSS treatment, cells were washed with cold PBS, fixed in methanol (prechilled at −20 °C) for 3 min and then incubated with rabbit polyclonal anti-ZO1 (D6L1E) rabbit monoclonal antibody (Cell Signaling Technology, 13663; 1:100) for 1 h. Cells were incubated with FITC-labeled secondary antibodies (AffiniPure goat anti-rabbit IgG (H + L), Jackson, 111-095-003; 1:50) for 1 h. DAPI (Invitrogen, D3571) was used to label DNA in nuclei. Slides were mounted on glass slides using the Prolong Gold antifade reagent (Invitrogen, P36970) and visualized using a confocal fluorescence microscope (Leica TCS SP8 MP Multiphoton Microscope, Leica Microsystem). Each figure (×68 magnification) represents eight fields captured for each coverslip. The mean fluorescence intensity of each figure was analyzed using ImageJ version 1.54g (National Institutes of Health).
BMDC culture and qPCR assay
Bone marrow cells were flushed from the femurs and tibiae of C57BL/6J mice with 0.5% BSA and 2 mM EDTA pH 8.0 in 1× PBS, passed through a nylon mesh and treated with ACK lysing buffer (Life Technologies, A10492-01) for 2 min. The cells were then washed and cultured in nonadherent Petri dishes with DMEM/F12 supplemented with GlutaMAX, 10% heat-shocked FBS (Gibco, A56697-01), 1% penicillin–streptomycin (Life Technologies, 15140122), 1% sodium pyruvate (Life Technologies, 11360070), 1% HEPES (Life Technologies, 15630106), 1% MEM nonessential amino acids (Life Technologies, 11140050) and 20 ng ml−1 granulocyte–macrophage colony-stimulating factor (GM-CSF; StemCell Technologies, 78017). The medium was replaced on days 3 and 5 with recombinant mouse GM-CSF to induce differentiation. By day 5, over 90% of the cells expressed dendritic cell-specific markers, as confirmed by fluorescence-activated cell sorting (FACS). On day 6, DCs were replated into new 24-well plates and treated with ISM012-042 (1, 3 or 10 µM) or roxadustat (10 µM). Control and treated BMDCs were also stimulated with LPS (100 ng ml−1) for 24 h. Subsequently, RNA extraction was performed using a Tiangen RNA extraction kit (DP761), followed by reverse transcription (RT) with PrimeScript RT Master Mix (Takara, RR036A). RT–qPCR was conducted using PowerUp SYBR green mix (Applied Biosystem, A25742) on a QuantStudio 6 Flex real-time PCR system (Applied Biosystems), with mRNA levels normalized to Gapdh using the 2−ΔΔCt method. All reactions were performed in triplicate. Primer sequences are provided in Supplementary Table 11.
Animal studies
General toxicity
Ten SD rats per sex per group (purchased from Vital River Laboratory Animal Technology) were used in 28-day repeat-dose general toxicity studies. The rats were 7–9 weeks old and the body weights ranged from 165 to 202 g for females and 300 to 350 g for males at initiation of dosing. During the study, the rats had ad libitum access to food (Beijing/Tianjin Keao Xieli Feed, 22073213 and 22113213) and water. The rats were housed in a temperature-controlled (20–24 °C) with 40–70% humidity and a 12-h light–dark cycle. ISM012-042 was suspended in 0.5% (v/v) Tween 80 with 0.5% (w/v) carboxymethyl cellulose (CMC)-32 solution at concentrations to provide a 0, 100, 300 or 1,000 mg kg−1 dose in a 10 ml kg−1 dose volume by oral gavage daily for 28 days. The toxicity assessment covered viability, clinical signs, body weights, food consumption, ophthalmologic examinations, clinical pathology (hematology, coagulation parameters, serum chemistry and urinalysis), gross necropsy observations, organ weight and histopathology. The protocol and any amendments or procedures involving the care or use of animals on this study were reviewed and approved (assurance identification number SZ20221201-Rats-A) by the WuXi AppTec IACUC before the initiation of such procedures.
Mice and chemically induced colitis
For the TNBS colitis model, female BALB/c mice aged 8 weeks (body weight: ~19–21 g) were purchased from Lingchang Biotech and were bred in specific pathogen-free individually ventilated cages in a temperature-controlled (20 ± 2 °C) room with 40–70% humidity and a 12-h light–dark cycle. Chow pellets and tap water were available ad libitum. The mice were acclimated at the animal facility for at least 3 days before the experiments. On day 0, mice were anesthetized and received a 100-μl rectal instillation of either vehicle (50% ethanol) or TNBS (2% TNBS in 50% ethanol). Daily treatment with compound or vehicle (0.5% CMC–Na plus 2% Tween 80) by oral gavage occurred on days −1 (prevention mode) or day 2 (therapeutic mode) through day 7.
For the oxazolone colitis model, female BALB/c mice aged 8 weeks (body weight: ~18–20 g) were purchased from Charles River Laboratories and bred in specific pathogen-free individually ventilated cages. On day −5, mice were presensitized with 200 μl of 2% oxazolone dissolved in a 4:1 mixture of acetone and olive oil on a 4-cm2 field of the shaved back, followed by intracolonic injection of 100 μl of either oxazolone (1.5% oxazolone dissolved in 50% ethanol) or vehicle (50% ethanol) on day 0. Before intracolonic injection, mice were anesthetized with 0.3 ml of avertin (1.25%) and maintained in the head-down position for 2 min following intracolonic administration. All experimental protocols were approved (assurance identification numbers IM01-005-2021v1.0 and IM01-SH003-2023v1.1) by Wuxi AppTec IACUC.
Evaluation of colitis severity
Body weight and DAI soring were recorded daily to assess colitis severity. DAI score was the sum of the weight loss, stool and bleeding subscores. A blinded scoring system was used to determine colitis severity. The DAI scorer, blinded to the group information and animal identifier, was responsible for the stool consistency and bleeding evaluations. DAI scoring standards are presented in Supplementary Table 12.
FOB test
If no blood was visible with the naked eye, the fecal occult blood (FOB) test was performed with an FOB test strip (the improved pyramidon method). The FOB score (0–2) was interpreted as follows: 0, no color appeared after 2 min; 1, a dimmed color appeared after 1–2 min; 2, a deep color appeared after 1–2 min.
Intestinal permeability assay
FITC–dextran (molecular weight, 4,000; 600 mg kg−1; Sigma-Aldrich, 46944) was administered by oral gavage after 4–6 h of fasting. Then, 4 h after dosing with FITC–dextran, mice were anesthetized and blood was sampled in K3 EDTA-coated tubes. Plasma was isolated by centrifugation at 2,000g for 10 min at 4 °C. FITC–dextran concentrations in plasma were analyzed in duplicate using a spectrophotometer with an excitation wavelength of 485 nm and emission wavelength of 535 nm. Baseline blood plasma fluorescence was determined in mice after oral gavage with water and subtracted from fluorescence obtained after FITC–dextran gavage. FITC–dextran concentrations were determined from standard curves generated by serial dilutions of FITC–dextran.
RNA extraction and real-time qPCR
Whole-colon tissue from each mouse was snap-frozen in liquid nitrogen and stored at −80 °C until analysis. Samples were homogenized with a Qiagen TissueLyser LT in TRI reagent and total RNA was purified using the RNeasy mini kit (Qiagen). RNA was converted into complementary DNA using the FastKing RT kit (Tiangen) according to the manufacturer’s instructions. qPCR was performed on a QuantStudio 7 Flex real-time PCR system (Applied Biosystems) using SYBR green master mix (Applied Biosystems). The relative expression was normalized to Gapdh using the 2−ΔΔCt method. Primers sequences are listed in Supplementary Table 11.
Flow cytometry
Lamina propria lymphocytes were isolated from the intestines of TNBS mouse models and subjected to antibody staining and flow cytometry as previously described25. Cell suspensions were washed with 1× PBS and stained for live–dead markers and surface antibodies on ice for 30 min, followed by a wash with FACS buffer (0.5% BSA and 2 mM EDTA in 1× PBS). Cells were stimulated with a cell stimulation cocktail plus transporter inhibitors for 4 h before staining to detect cytokines. Intracellular staining was performed using an intracellular labeling kit, with cells fixed, permeabilized and then stained with antibodies in 1× perm washing buffer for 30 min at 4 °C. Intracellular antibodies included interferon-γ, IL-17A, forkhead box P3, TNF and retinoic acid-related orphan receptor-γt, while all other antibodies were used for surface staining. All antibodies were used at dilution of 1:50. Sample collection and data analysis were performed using a BD Fortessa flow cytometry analyzer and FlowJo software (version 10). Details of antibodies and reagents are provided in Supplementary Table 13.
Lymphocytes were initially selected on the basis of forward scatter (FSC) and side scatter (SSC) and doublets were excluded using FSC-A/FSC-W and SSC-A/SSC-H plots. Dead cells were excluded by gating on the FVS620low population. CD45+ cells were then selected to identify pan-leukocytes.
Histopathology
The colon was removed and its length was measured when it was in a relaxed position without stretching; it was weighed after feces removal. The colon was opened longitudinally, washed thoroughly with PBS, rolled from the proximal to distal end, neutralized and fixed in paraformaldehyde, followed by staining with hematoxylin and eosin. Slides were scored by pathologists blinded to the animal identifier. The histological features that were assessed are listed in Supplementary Table 14. A total histological severity score, ranging from 0 to 11, was obtained by summing the eight-item scores.
HIF1α immunohistochemistry and quantification
Colon Swiss rolls embedded in formalin-fixed paraffin-embedded tissue block were sectioned into 4-μm-thick slides. The slides were deparaffinized, rehydrated and incubated in epitope retrieval solution 1 (pH 6) for 20 min at 100 °C. The primary antibody to HIF1α clone E1V6A (1:400; Cell Signaling Technologies, #48085) was incubated for 30 min at room temperature followed by bond polymer refine detection (Leica Biosystems, DS9800). Slides were digitized using Aperio VERSA 8 (Leica), and image acquisition was performed using Aperio ImageScope version 12.4 (Leica). Eight nonoverlapping fields of view per each colon Swiss-roll section were analyzed by HALO software (Indica Labs). For each colon Swiss-roll section, the percentage of HIF1α and nucleus double-stained counts in total cell counts (12,000–14,000 cells counted from eight fields of view) were quantitated for the mucosa layer at the midsegment to the distal end of the colon roll at ×200 magnification.
Cytokine and chemokine analysis
Whole-colon or half-colon tissue was cut longitudinally and homogenized and colonic cytokines were measured using the V-PLEX proinflammatory panel one mouse kit (Meso Scale Discovery, K15048D) and V-PLEX cytokine panel one mouse kit (Meso Scale Discovery, K15245D). Plasma EPO, VEGFA, IL-1β, IL-6 and TNF were measured using customized U-PLEX biomarker group 2 assays (Meso Scale Discovery, K152ADM).
Fecal biomarker measurement
Mice were placed in clean polycarbonate cages without bedding until they defecated or for 1 h, whichever came first. The samples were homogenized in radioimmunoprecipitation assay buffer or other lysis buffer containing protease inhibitor with a 1:5 (w/v) ratio after weighing followed by sonication and centrifugation at 13,800g for 10 min at 4 °C. Total protein concentration of supernatant from fecal samples was determined using a BCA protein assay kit (Thermo Fisher Scientific, 23225) for the final normalization. Calprotectin and LCN2 were detected using the respective ELISA kits (Elabscience, E-EL-M1143c; Beyotime, PN757).
RNA-seq data processing
Around 100 mg of colon tissue was collected from sham or TNBS-induced colitis mouse models treated with vehicle or ISM012-042 on day 2 after colitis induction. The mRNA isolation and library preparation for next-generation sequencing were conducted described above. Libraries were sequenced using the Illumina Novaseq. Raw sequencing data obtained were processed using the DRAGEN (version 4.0.3) pipeline, which aligned the reads against the National Center for Biotechnology Information (NCBI) RefSeq mouse genome (mm10) to generate count data. The quality of sequencing and alignments was assessed using MultiQC (version 1.12)26. Differential gene expression analysis was performed using the DESeq2 package (version 1.34.0) with R (version 4.1.2)27. Subsequently, the log2 fold changes derived were used for gene set enrichment analysis conducted with the fgsea package (version 1.20.0) with R (version 4.1.2)28 and the P value was adjusted using the Benjamini–Hochberg correction method. The enrichment analysis used gene sets from the Kyoto Encyclopedia of Genes and Genomes pathway database29.
Quantitative MS
Colonic mucosa obtained from TNBS-induced colitis models treated with GB004, ISM012-042 or vehicle were homogenized and assayed by tandem mass tag MS, as described previously30 and analyzed using the MASCOT engine (Matrix Science, version 2.2) embedded in Proteome Discoverer 1.4 software against the UniProt mouse database (17,097 sequences; downloaded on October 4, 2022) for identification and quantitation. Differentially expressed proteins were identified using a two-sided t-test in R (version 4.1.2).
Statistics
Experiments were repeated at least three times, with one representative dataset shown. Data are presented as the mean ± s.d. Statistical analyses were performed using GraphPad Prism versions 8.0 and 10.1. A P value < 0.05 was considered statistically significant. The Shapiro–Wilk test or Kolmogorov–Smirnov test was used to test the normality of data distribution. A two-sided, unpaired Mann–Whitney U-test was used for comparisons between two groups. One-way or two-way analyses of variance (ANOVAs) with Tukey’s or Dunnett’s multiple-comparisons tests were used with the P value adjusted for multiple comparisons as indicated in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.