Sequence representation
To apply the diffusion framework in sequence space, a continuous representation of the categorical sequence data is needed. To implement this, we represented the sequence, x0, with dimensions L×20 where L corresponds to the protein length with 20 possibilities for each amino acid type. This takes the form of a one-hot encoded vector that is centered at zero by multiplying the L×20 tensor by 2 and subtracting 1. Each logit within the tensor is a real number, with higher values corresponding to a higher probability for that specific amino acid at that position. With this representation, we noise x0 to obtain xt with the below equation following the Ho et al.8 formulation for a standard forward process sampling from Gaussian noise with mean at 0 and standard deviation of 1.
$$q({{\bf{x}}}_{t}|{{\bf{x}}}_{0})={\mathscr{N}}({{\bf{x}}}_{t};\sqrt{{\bar{\alpha }}_{t}}{{\bf{x}}}_{0},(1-{\bar{\alpha }}_{t}){\bf{I}})$$
A critical part of the forward diffusion process is selecting the noising schedule. Determining the correct bin of a categorical distribution is trivial at low timesteps by argmaxing the input sequence. Therefore, more noise should be present at low timesteps to increase the difficulty of the task during training. The square root noise schedule10 satisfies this requirement and was employed in this study.
Training
To train the model, we began by sampling t uniformly from [0,T], where t = 0 is an un-noised sequence and t = T is pure Gaussian noise. We then noise x0 to xt with equation (1) and tasked the model to predict the un-noised sequence x0 and its corresponding structure y. The timestep feature was added to the sequence template passed to the model. We applied a categorical cross-entropy loss to x0 and structure losses to y (FAPE, bond angle, bond length, distogram, lddt). An additional KL loss10 was applied to the calculated xt−1, as previously demonstrated to stabilize training of discrete diffusion models10. Self-conditioning16 was implemented to allow the model to condition on the previous x0 prediction and the back-calculated xt−1 during both training and inference. To self-condition in practice, the model was used with gradients turned off to first predict x0 from xt+1, which was then passed in as a sequence template to the model. During training, RoseTTAFold was allowed 1–3 uniformly sampled ‘recycle’ steps to refine structure predictions via multiple passes through the model53. Pseudo training and inference code is available in the Supplementary Information (Algorithms S1 and S2). In later training iterations, secondary structure conditioning was provided to the model by concatenating a tensor representing DSSP features onto the sequence template. These features were provided 25% of the time and masked uniformly between 0% and 90% when provided.
Along with the standard diffusion task (40% of the time), the model was also challenged with structure prediction (seq2str) and fixed backbone sequence design (30% of the time each). Incorporating these additional tasks during training helped maintain the agreement of sequence–structure pairs diffused by the model. Training examples were conditioned on sequence or structure by either unmasking 1–4 spans of residues, each 4–8 amino acids in length to simulate motif scaffolding, or unmasking randomly selected residues for the model to scaffold as an active site scaffolding problem. Unmasked structure conditioning information was supplied to the input for RoseTTAFold as templates in the 1D sequence track as well as the 2D and 3D structural information tracks.
Inference
During inference starting from xt, the model predicts x0 and simultaneously decodes it to y. x0 is then back-calculated to xt−1 with equation (1) and passed through the network with the previously predicted x0 to apply self-conditioning. Benchmarking against conditioning on xt, as done in Ho et al.8 with the below equation, shows that this approach performs better (Supplementary Fig. 3c), as seen in other categorical diffusion methods10,17.
$$q({{\bf{x}}}_{t-1}|{{\bf{x}}}_{t},{{\bf{x}}}_{0})={\mathscr{N}}({{\bf{x}}}_{t-1};{\tilde{{\boldsymbol{\mu }}}}_{t}({{\bf{x}}}_{t},{{\bf{x}}}_{0}),{\tilde{\beta }}_{t}{\bf{I}}),$$
where \({\tilde{{\boldsymbol{\mu }}}}_{t}({{\bf{x}}}_{t},{{\bf{x}}}_{0}):=\frac{\sqrt{{\bar{\alpha }}_{t-1}}{\beta }_{t}}{1-{\bar{\alpha }}_{t}}{{\bf{x}}}_{0}+\frac{\sqrt{{\alpha }_{t}}(1-{\bar{\alpha }}_{t-1})}{1-{\bar{\alpha }}_{t}}{{\bf{x}}}_{t}\) and \({\tilde{\beta }}_{t}:=\frac{1-{\bar{\alpha }}_{t-1}}{1-{\bar{\alpha }}_{t}}{\beta }_{t}\)
This is done for T steps, but T can be varied and does not have to be what was used during training (inference time for fixed T can be found in Supplementary Table 2). The model finds solutions to some problems in as few as 10 steps (Fig. 1c). Furthermore, clamping the model’s output logits from −3,3 gives better agreement with AF2 predictions (Supplementary Fig. 3b). xt−1 is sampled from either a zero-mean normal distribution or a non-Bayesian Gaussian mixture distribution with equal mixing probabilities. For the non-Bayesian Gaussian mixture models, we defined a mixture with two normals centered at [−1, 1] (GMM2) and a mixture with three normals centered at [−1, 0, 1] (GMM3).
Unconditional protein generation
Unconditionally generated proteins were assessed against a set of 1,000 native proteins with a length deviating up to five residues randomly sampled from the RCSB54 database. For experimental verification, proteins ranging from 70 to 80 amino acids in length with no conditioning information were generated in 25 steps. Designs were filtered by AF2 pLDDT > 90 and AF2 RMSD to design < 2 Å for ordering final constructs. Additionally, proteins with high model confidence but moderate AF2 confidence were ordered by filtering on design pLDDT > 90, AF2 pLDDT < 80 and AF2 RMSD to design < 5 Å.
Compositionally biased protein generation
Proteins ranging from 70 to 80 amino acids in length with an amino acid compositional potential were generated in 25 steps. Designs were filtered by AF2 pLDDT > 90, AF2 RMSD to design < 2 Å and SAP score55 < 30. The top 10–22 designs were ordered for each upweighted amino acid type (tryptophan, cysteine, valine, histidine and methionine). Pseudocode for the implementation of the amino acid compositional potential is provided in the supplements (Algorithm S3).
Charge biased protein generation
Proteins of 50 amino acids in length with charge potentials applied were generated in 25 steps with charge conditioning information. The ground truth charge for each protein was calculated at pH 7.4 by using the Henderson–Hasselbach equation.
Hydrophobic biased protein generation
Proteins of 50 amino acids in length with hydrophobic potentials applied were generated in 25 steps with hydrophobicity conditioning information. The ground truth hydropathy index for each design was calculated by summing the hydropathy index for each residue and dividing by the sequence length56.
DSSP guidance
For constructing the DSSP features, we calculated each training example’s DSSP based on the structure with helix, strand, loop and masked labels57. During training, the calculated per-residue secondary structure features were appended to RoseTTAFold’s 1D features and were one-hot encoded for 25% or 50% of the time and masked for 30% or 80% of the time. During inference, DSSP features are appended to the 1D features as necessary and masked when not. Secondary structure representations were input to the model as follows: H, helix; E, sheet; L, loop; X, masked.
Repeat protein generation
Repeat proteins ranging from 125 to 150 amino acids in length were generated in 50 steps with and without DSSP conditioning information. Designed proteins contained five repeat units using one of the following DSSP strings, where X represents mask, E represents strand and H represents helix:‘XXXXEEEEEXXXXXXXXXXXXXXXHHHHHXXXX’, ‘XXXXEEEEEXXXXXHHHHHXXXXXEEEEEXXXX’, ‘XXXXHHHHHXXXXXEEEEEXXXXXHHHHHXXXX’, ‘XXXXHHHHHXXLXXHHHHHXXLXXHHHHHXXXX’, ‘XXXXEEEEEXXHXXEEEEEXXHXXEEEEEXXXX’, ‘XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX’, ‘XXXXEXXXXXXXXXEXXXXXXXXXEXXXXXXXX’. Designs were filtered on AF2 pLDDT > 80 and AF2 RMSD to design < 2 Å.
Caging bioactive peptides
Proteins of 155 amino acids in length were generated in 25 steps with spans of helical DSSP conditioning to encourage the model to generate helical bundles and cleavage loops. Additional DSSP features were provided to scaffold the furin protease cleavage site ‘GRRKR’. The sequence for melittin was provided without DSSP conditioning as N-terminal 26 amino acids ‘GIGAVLKVLTTGLPALISWIKRKRQQ’. Designs were filtered by AF2 pLDDT > 85, AF2 RMSD to design < 2 Å and SAP scores < 40.
Scaffolding barcode peptide sequences
Proteins of 100 amino acids in length were generated in 50 steps with barcodes being set as fixed regions on the C-terminus. Designs were filtered by AF2 pLDDT > 85 and AF2 RMSD to design < 2 Å.
Multistate guidance
Parent and child protein pairings were generated in 25 steps using secondary structure features ‘XHHHHHHHHHHHHHLLLHHHHHHHHHHHHHLLLEEEEEELLLEEEEEXXXXXXXEEEEELLLE EEEEELLLHHHHHHHHHHHHHLLLHHHHHHHHHHHHHX’ for parent, ‘XHHHHHHHHHHHHHLLLHHHHHHHHHHHHHLLLHHHHHHHHHHHHHHHHHHHX’ for child A and ‘XHHHHHHHHHHHHHHLLLHHHHHHHHHHHHHLLLHHHHHHHHHHHHHX’ for child B. A mixing coefficient of 0.25 was used to combine parent and child sequences together at each step. Multistate design pseudocode implementation is available in the supplements (Algorithm S4).
Partial noising
To promote further exploration of known active sequence subspaces, we implemented partial noising during design trajectories. This was done by introducing a temperature parameter, λ ∈ (0 to 1], to PG that, at the beginning of a trajectory, sets t to round (T*λ) rather than T (complete noising). This forced the model to do local exploration of the partially noisy sequence subpace. This approach was done in conjunction with normal design trajectories in ‘multistate guidance’ with λ parameters iterated through [0.2,0.3,0.5,0.8] starting with passing AF2 design sequences.
Iterative guidance
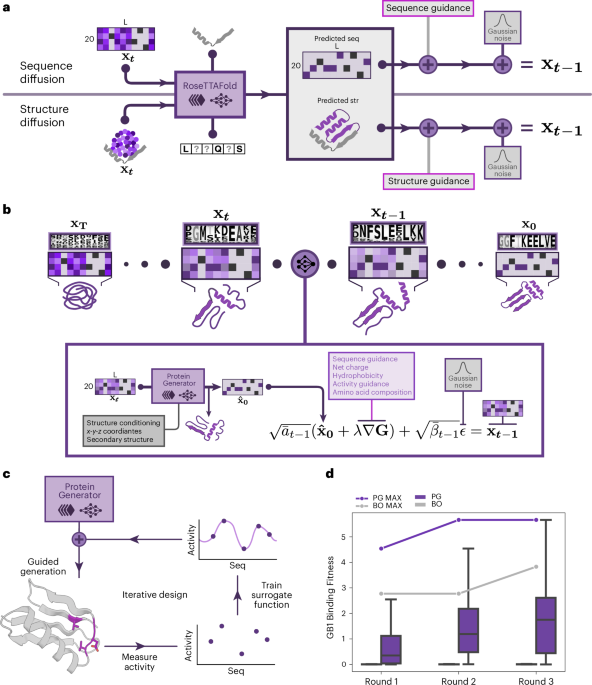
Iterative guidance was employed as described in Fig. 1c. In short, the experimental or in silico characterization of designs is collected to train a classifier, which is used to generate further designs with guidance. In an iterative process, classifiers trained on designs sampled in a preceding round inform network sampling. We investigated the nearly complete fitness landscape of the V39, D40, G41 and V54 amino acid sites of GB1, which is the binding domain of protein G, an immunoglobulin binding protein found in streptococcal bacteria, as provided in the FLIP58 paper. Three rounds of iterative design were conducted with a batch size of 96.
We performed diffusion on the four mutation sites in the GB1 protein while providing guidance using a scale of 2.0 in the second-round and third-round design processes. In the first round, designs are directly sampled from our protein generator. For each round, a vanilla MLP (two layers, rectified linear unit (ReLU) activation and dropout of 0.25 after the first layer) is trained on the designs generated in the preceding round using a fitness equal to 1 as the classifier boundary. For comparison, batched Bayesian optimization was performed for three rounds using the sequences generated in the first round of the iterative guidance as the initial dataset. For Bayesian optimization, we used Gaussian processes with the radial basis function (RBF) kernel for the surrogate function, and we used the Monte Carlo–based batch upper confidence bound (qUCB) acquisition59 function with λ = 0.5.
Sequence and structure quality metrics
All designs used in the ESMFold21 benchmarks were modeled by using the following curl command: [curl -X POST–data ‘SEQUENCE’] (https://api.esmatlas.com/foldSequence/v1/pdb/). All designs that were used in AF2 benchmarks and ordered for experimental characterization were predicted in single-sequence structure prediction mode with model 4. Pairwise backbone RMSDs between the design model and AF2 model were calculated for each design. Sequence quality metrics measured by ESM21 (pseudo) perplexity were calculated with model: esm2_t33_650M_UR50D.
Sequence identity calculations
Blast alignment was used to examine sequence alignment and similarity using query coverage >90% and target coverage >50%. Alignment to natives was done against UniRef90 (Supplementary Fig. 27).
Plasmid construction
Protein designs were cloned into plasmids as in Watson et al.11. In brief, designs were ordered as synthetic genes (eBlocks, Integrated DNA Technologies) with BsaI overhangs compatible with a ccdB-encoding expression vector, LM0627 (ref. 7). Genes cloned into LM0627 result in the following sequence: MSG-design-GSGSHHWGSTHHHHHH (SNAC cleavage tag and 6×His affinity tag are indicated). We used the NEBridge Golden Gate Assembly Kit (New England Biolabs) with a total reaction volume of 5 μl and a ratio of 1:2 by mass of LM0627 plasmid DNA to design. We then incubated the reaction mixture at 37 C for 30 min, halted the reaction by incubating the reaction mixture at 60 °C for 5 min and transformed 1 μl of the reaction mixture into 6 μl of BL21 competent cells (New England Biolabs). After heat shock and recovery in SOC media, transformed BL21 cells were grown overnight in 1.0 ml of LB from which glycerol stock was created and small-scale expression cultures were inoculated.
1-ml-scale protein purification
Initially, proteins were expressed with small-scale expression screens as previously reported7 with small adaptations. In brief, designs were inoculated with 100 µl of overnight growths and 900 µl of auto-induction media (sterile-filtered TBII media supplemented with 50 µg ml−1 kanamycin, 2 mM MgSO4, 1 × 5,052) in deep-well 96-well plates. Sixteen hours after inoculation, cells were harvested and lysed in lysis buffer (50 mM Tris-HCl (pH 8), 0.5 M NaCl, 30 mM imidazole supplemented with 1× BugBuster, 1 mM PMSF, 0.1 mg ml−1 lysozyme, 0.1 mg ml−1 DNase). Clarified lysates were added to a 50-μl bed of Ni-NTA agarose resin in a 96-well fritted plate equilibrated with wash buffer (50 mM Tris-HCl (pH 8), 0.5 M NaCl, 30 mM imidazole). After sample application and flowthrough, the resin was washed three times with wash buffer, and samples were eluted in 200 μl of elution buffer (50 mM Tris-HCl (pH 8), 0.3 M NaCl, 0.5 M imidazole, 5 mM EDTA (pH 8)). All eluates were sterile filtered with a 96-well 0.22-μm filter plate (Agilent, 203940-100) before SEC. Protein designs were then screened via SEC using an ÄKTA FPLC outfitted with an autosampler capable of running samples from a 96-well source plate. Samples were run on a Superdex S75 Increase 5/150 GL column (Cytiva, 29148722; 3,000–70,000-Da separation range) in a running buffer (20 mM Tris (pH 8), 150 mM NaCl). To improve peak resolution, the SEC column was connected directly in line from the autosampler to the UV detector. Then, 0.25-ml fractions were collected from each run. Absorption spectra were collected by an ÄKTA U9-M at 230 nm and 280 nm.
50-ml-scale protein purification
Proteins selected for further downstream characterization were expressed in 50 ml of auto-induction media60. Sixteen hours after inoculation, cells were harvested and lysed in lysis buffer (50 mM Tris-HCl (pH 8), 0.5 M NaCl, 30 mM imidazole, 1 mM PMSF, 0.1 mg ml−1 lysozyme, 0.1 mg ml−1 DNase) through sonication. Clarified lysates were added to a 2-ml bed of Ni-NTA agarose resin in a 20-ml column (Bio-Rad, 7321010) equilibrated with wash buffer (50 mM Tris-HCl (pH 8), 0.5 M NaCl, 30 mM imidazole). After sample application and flowthrough, the resin was washed three times with 10 ml of wash buffer, and samples were eluted in 2 ml of elution buffer (50 mM Tris-HCl (pH 8), 0.5 M NaCl, 200 mM imidazole). All eluates were sterile filtered with a 3-ml 0.22-µm filter plate before SEC. Protein designs were then screened via SEC using an ÄKTA FPLC outfitted with an autosampler capable of running samples from a 96-well source plate. Samples were run on a Superdex S75 Increase 10/300 GL column (Cytiva, 29148721; 3,000–70,000-Da separation range) in a running buffer (20 mM Tris (pH 8), 150 mM NaCl). Then, 1-ml fractions were collected from each run. Absorption spectra were collected by the ÄKTA U9-M at 230 nm and 280 nm.
0.5-L-scale protein purification and SNAC cleavage
The best expressing proteins used in high-resolution structural studies were selected for further scale-up and SNAC cleavage61. Proteins were expressed in Studiers M2 autoinduction media with 50 µg ml−1 kanamycin. Pre-cultures were grown overnight. Cultures were inoculated with 10 ml of pre-culture and grown at 37 °C for 4 h before lowering temperature to 22 °C for 14 h, and cultures were inoculated with 10 ml of pre-culture. Cells were pelleted at 4,000g for 10 min, after which the supernatant was discarded. Pellets were resuspended in 30 ml of lysis buffer (100 mM Tris HC (pH 8), 100 mM NaCl, 400 mM imidazole, 1 mM PMSF, 1 mM DNase). Cell suspensions were lysed by sonication for 7.5 min (10 s on, 10 s off) at 80% amplitude using a Qsonica four-prong sonicator. The lysate was clarified at 14,000g for 30 min. The His-tagged proteins were batch bound for 1 h to 8 ml of Ni-NTA resin (Qiagen) and washed with 10 ml of lysis buffer and 30 ml of high-salt wash buffer (25 mM Tris HCl (pH 8), 1 M NaCl, 40 mM imidazole) and then 10 ml of SNAC cleavage buffer (100 mM CHES, 100 mM acetone oxime, 100 mM NaCl, 500 mM GnCl (pH 8.6)). Next, 40 ml of SNAC cleavage buffer and 80 µl of 1 M NiCl2 were added, and columns were closed and shaken on a nutator for 12 h to cleave. After cleavage, the flowthrough was collected and concentrated before further purification by SEC/FPLC as described above.
Cysteine bias protein expression
Proteins guided toward high cysteine content were transformed into and expressed in Rosetta-gami B(DE3) competent cells (Novagen, 71137). The 1-ml and 50-ml scale protein purification protocols were otherwise followed.
CD
CD spectra were collected on a Jasco J-1500 CD spectrometer with 1-nm bandwidth, 50-nm permanent scan rate and data integration time of 4 s per read. Sample cuvettes stored in 2% Hellmanex (Hellma, 9-307-011-4-507) were washed with deionized water, 2% Hellmanex, deionized water and then 20% ethanol, after which 300 µl of SEC-purified protein was added for CD spectra measurements. Thermal melts were performed in 10° intervals between 25 °C and 95 °C.
Mass spectrometry
To identify the molecular mass of each protein, intact mass spectra were obtained via reverse-phase liquid chromatography–mass spectrometry (LC–MS) on an Agilent G6230B TOF on an AdvanceBio RP-Desalting column and subsequently deconvoluted by way of BioConfirm using a total entropy algorithm. Disulfide formation was determined by injecting protein at 1.5 mg ml−1 in the presence and absence of 50 mM TCEP-HCl (Millipore Sigma, 646547-10X1ML) and detecting the mass shift.
Disulfide bond quantification
To measure the number of cysteines via alkylation, proteins at 1.5 mg ml−1 in SEC running buffer (20 mM Tris (pH 8), 150 mM NaCl) were incubated in 50 mM TCEP-HCl at 50 °C for 1 h to reduce disulfide bonds. Simultaneously, an equal amount of protein in SEC running buffer was heated to 50 °C without 10 mM TCEP to maintain formed disulfides. Iodoacetamide (Millipore Sigma, I1149) was added to both conditions to a final concentration of 10 mM and incubated away from light at room temperature for 30 min to alkylate unpaired cysteines. To identify the molecular mass and alkylations status of each protein, intact mass spectra were obtained via reverse-phase LC–MS on the Agilent G6230B TOF on an AdvanceBio RP-Desalting column and subsequently deconvoluted by way of BioConfirm using a total entropy algorithm.
Barcode extraction and liquid chromatography–tandem mass spectrometry
Ni-NTA eluate of the 84-design pool was subjected to SEC with deep fractionation (0.25-ml fractions). From every other fraction, 100 µl was added to fresh wells in a 96-well plate, and fractions were subjected to cleavage in 100 µl of Lys-C buffer (8 M urea, 100 mM Tris HCl (pH 8)) plus 1 µg of endoproteinase LysC (New England Biolabs, P8109S), as previously described. After hexaHis-tagged barcode pulldown with magnetic His-pulldown beads (Thermo Fisher Scientific, 10103D) and subsequent trypsin (New England Biolabs, P8101S) digest to free barcodes, barcodes were diluted 50% in 0.1% trifluoroacetic acid (TFA). Barcode pools corresponding to SEC fractions were separated by hydrophobicity using a previously described tandem guard column–analytical column setup. The guard column was packed to 2 cm with 5 µm of silica (ReproSil-Pur 120 C18Aq, ESI Source Solutions, r15.aq.0001), whereas the analytical column was packed to 14 cm with 1.9 µm of silica (ReproSil-Pur 120 C18Aq, ESI Source Solutions, r119.aq.0001). Peptides were detected using a previously described data independent acquisition (DIA) protocol on a Orbitrap Fusion Lumos Tribrid (Thermo Fisher Scientific) at the UW Proteomic Resource (UWPR).
Solution NMR
Recombinant plasmid DNA (~100 ng) containing synthetic genes encoding for child A, child B and parent for several design families were separately transformed in E. coli BL21(DE3) cells. Colonies were grown under kanamycin selection on LB agar media for 16 h. Toward preparation of uniformly 15N-labeled proteins, a streak of colonies was resuspended in 60 ml of 1× M9 minimal media62 and grown overnight at 37 °C/225 r.p.m., and the inoculum was used to initiate a 1-L 1× minimal media culture supplemented with kanamycin and 15N ammonium chloride (Cambridge Isotope Laboratories, NLM-467) as the nitrogen source. For 15N/13C-labeled proteins, 13C-labeled glucose (Cambridge Isotope Laboratories, CLM-1396) was used as the carbon source. Cultures were incubated at 37 °C/225 r.p.m. until the optical density at 600 nm (OD600) reached 0.6 and then induced with 1 mM IPTG and grown at 37 °C/225 r.p.m. for 6 h. Cultures were harvested by centrifugation (6,000g, 15 min, 4 °C), and cell pellets were resuspended with wash buffer (300 mM NaCl, 10 mM imidazole, 50 mM Tris (pH 8)). Cells were lysed by sonication on ice. The lysate was clarified by centrifugation at 10,000g for 20 min at 4 °C. The supernatant was loaded onto a 5-ml His-Trap Ni-NTA column. The column was washed extensively with wash buffer, and protein was eluted using a linear gradient from 0% to 100% elution buffer (300 mM NaCl, 500 mM imidazole, 50 mM Tris (pH 8)). Fractions containing protein were pooled and further purified by SEC on a Superdex 200 Increase 10/300 GL column in NMR buffer (100 mM NaCl, 20 mM sodium phosphate (pH 6.5)). All designs were purified into batch-matched NMR buffer and then concentrated to 300 μl in 3-kDa Amicon concentrators. The purity of eluent fractions was confirmed to be greater than 95% by SDS-PAGE. Protein concentrations were measured by NanoDrop spectrophotometer at 280 nm with extinction coefficient predicted by ExPASY ProtParam. 2D 1H-15N amide HSQC spectra (Bruker pulse sequence hsqcetf3gpsi) were acquired using standard parameters at a 1H field of 800 MHz at 37 °C with recycle delay (d1) set to 1.2 s, sweep width of 30 ppm and acquisition time of 60 ms and number of scans ranging from 8 to 64 on a Bruker AVIIIHD-800 spectrometer equipped with a 3-mm TCI cryoprobe. All data were processed in NMRPipe63 and analyzed in NMRFAM-SPARKY64.
Uniformly double-labeled 15N/13C design proteins were prepared in NMR buffer as described above at final concentrations of 200 μM to 1,400 μM. Backbone HN, N, Cα, C13 and CO resonances were assigned using sequential assignment strategies65 via standard triple-resonance experiments with non-uniform sampling with 20% Poisson gap sampling schedule and were reconstructed with istHMS10 (http://gwagner.med.harvard.edu/intranet/hmsIST/). The following experiments were recorded: 3D HNCA (Bruker pulse sequence hncagpwg3d), 3D HNCO (Bruker pulse sequence hncogpwg3d), 3D HNCACB (Bruker pulse sequence hncacbgpwg3d) and 3D CBCACONH (Bruker pulse sequence cbcaconhgpwg3d). Acquisition times were 92 ms in 1H, 15 ms in 15N, 20 ms in 13CO and 10/5 ms in 13Cα/C13. Recycle delay was set to 1 s in all experiments, which were recorded at a 1H field of 800 MHz at 37 °C. To obtain through-space restraints for structure calculation, 3D amide–amide NOESY experiments (3D SOFAST HNHAro-NHN) were collected with 8–16 scans, 0.6-s recycle delay and 350-ms mixing time66. Nuclear Overhauser effect (NOE) cross-peaks were assigned manually in NMRFAM-SPARKY. NMR peak assignments were used by TALOS-N44 to determine secondary structure information, random coil index order parameter predictions and dihedral angle restraints toward structure calculation. Structure calculations were set up with automated Python scripts using CS-Rosetta67,68. We first used TALOS-N to determine psi and phi dihedral angles, and we used protein design sequences and assigned chemical shift values to pick fragments of amino acid lengths 3 and 9. We then used the protein sequence, 3mer/9mer fragments, backbone chemical shifts and amide–amide NOEs as input for the abrelax CS-Rosetta protocol (Rosetta version 3.8 and CS-Rosetta Toolbox version 3.3). From the 30,000 decoys calculated, the 10 lowest energy models were selected to represent the final NMR ensemble structure. The structure calculation was considered converged because the lowest energy models clustered within less than 2 Å from the model with the lowest energy. Final structures were validated with MolProbity.
1HN and 15N ACS values were determined from peaks in 2D 1H-15N HSQC spectra using the following equation43,69:
$${{\rm{ACS}}}_{{\rm{i}}}=\frac{1}{N}\sum _{K=1,M}{\omega }_{k}$$
where i = 1HN and 15N atoms; N is the total number of peaks picked in the HSQC spectrum; M is the total number of residues in the protein sequence; and wk is the chemical shift of the k-th resonance.
Reference 1HN and 15N ACS values for primarily α-helical proteins and primary 13-sheet proteins were taken from previous reports by Mielke et al.43,69.
Furin cleavage
To cleave designed proteins, 5 U of furin protease (New England Biolabs, P8077S) was combined with 30 µM design in enzyme buffer (20 mM HEPES, 1 mM CaCl2, 0.2 mM 13-mercaptoethanol) and incubated at 25 °C for 16 h. Cleavage reaction was used for SDS-PAGE (Any kDTM Mini-PROTEAN TGXTM Precast Protein Gels) with protein standards (Precision Plus Protein Dual Color Standards).
Blood cell lysis assay
Hemolysis assay was performed as described previously.70 Single-donor washed human RBCs (Innovative Research, IWB3ALS40ML) were washed three times by spinning blood at 500g for 5 min and discarding supernatant until supernatant appears clear. PBS was used to resuspend the RBCs at 10% hematocrit (v/v). Blood cell lysis was carried out in a 96-well plate at a final hematocrit of 2.5%. Negative controls include PBS, cleavage buffer and cleavage buffer with 0.5 U of furin. Positive controls include 2% Triton-X-100 (Sigma-Aldrich, 9036-19-5) and 15 µM melittin (GenScript, RP20415). Designed proteins were diluted to 15 µM with PBS. Washed RBCs were added to each well and incubated at 37 °C for 1 h, after which the reaction plate was spun down at 500g for 5 min. Supernatant from the reaction plate was transferred to a 96-well clear-bottom microplate (Corning, 3598). Absorbance was measured at 450 nm on an Agilent BioTek Epoch 2 TSC microplate reader.
Crystallography
All crystallization experiments were conducted using the sitting drop vapor diffusion method. Crystallization trials were set up in 200-nl drops using the 96-well plate format at 20 °C.
Crystallization plates were set up using a mosquito LCP from SPT Labtech and then imaged using UVEX microscopes and UVEX PS-256 from JAN Scientific. Diffraction quality crystals formed in a mixture of 0.1 M PCB buffer (pH 4) and 25% PEG 1500.
Diffraction data were collected at the National Synchrotron Light Source II. X-ray intensities and data reduction were evaluated and integrated using XDS71 and merged/scaled using Pointless/Aimless in the CCP4 program suite72. Structure determination and refinement starting phases were obtained by molecular replacement using Phaser73 using the designed model for the structures. After molecular replacement, the models were improved using phenix.autobuild74. Structures were refined in Phenix74. Model building was performed using Coot75. The final model was evaluated using MolProbity76. Data collection and refinement statistics are recorded in Table 1. Data deposition, atomic coordinates and structure factors reported in this paper have been deposited in the PDB with accession code 8VD6.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.